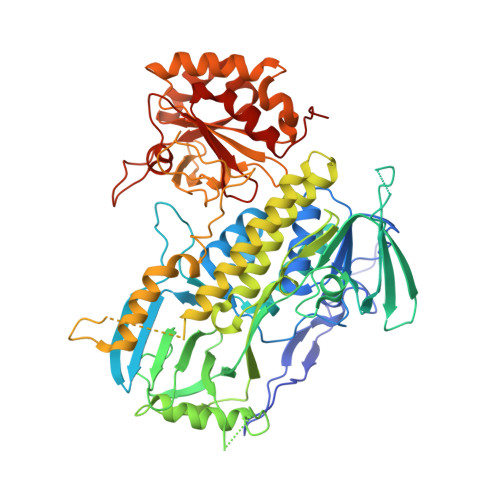
A flavin-monooxygenase catalyzing oxepinone formation and the complete biosynthesis of vibralactone.
Feng, K.N., Zhang, Y., Zhang, M., Yang, Y.L., Liu, J.K., Pan, L., Zeng, Y.(2023) Nat Commun 14: 3436-3436
- PubMed: 37301868
- DOI: https://doi.org/10.1038/s41467-023-39108-x
- Primary Citation of Related Structures:
7YJ0 - PubMed Abstract:
Oxepinone rings represent one of structurally unusual motifs of natural products and the biosynthesis of oxepinones is not fully understood. 1,5-Seco-vibralactone (3) features an oxepinone motif and is a stable metabolite isolated from mycelial cultures of the mushroom Boreostereum vibrans. Cyclization of 3 forms vibralactone (1) whose β-lactone-fused bicyclic core originates from 4-hydroxybenzoate, yet it remains elusive how 4-hydroxybenzoate is converted to 3 especially for the oxepinone ring construction in the biosynthesis of 1. In this work, using activity-guided fractionation together with proteomic analyses, we identify an NADPH/FAD-dependent monooxygenase VibO as the key enzyme performing a crucial ring-expansive oxygenation on the phenol ring to generate the oxepin-2-one structure of 3. The crystal structure of VibO reveals that it forms a dimeric phenol hydroxylase-like architecture featured with a unique substrate-binding pocket adjacent to the bound FAD. Computational modeling and solution studies provide insight into the likely VibO active site geometry, and suggest possible involvement of a flavin-C4a-OO(H) intermediate.
Organizational Affiliation:
State Key Laboratory of Phytochemistry and Plant Resources in West China and Yunnan Key Laboratory of Natural Medicinal Chemistry, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, 650201, China.