Structural analysis of the housecleaning nucleoside triphosphate pyrophosphohydrolase MazG from Mycobacterium tuberculosis.
Wang, S., Gao, B., Chen, A., Zhang, Z., Wang, S., Lv, L., Zhao, G., Li, J.(2023) Front Microbiol 14: 1137279-1137279
- PubMed: 36937295
- DOI: https://doi.org/10.3389/fmicb.2023.1137279
- Primary Citation of Related Structures:
7YH5 - PubMed Abstract:
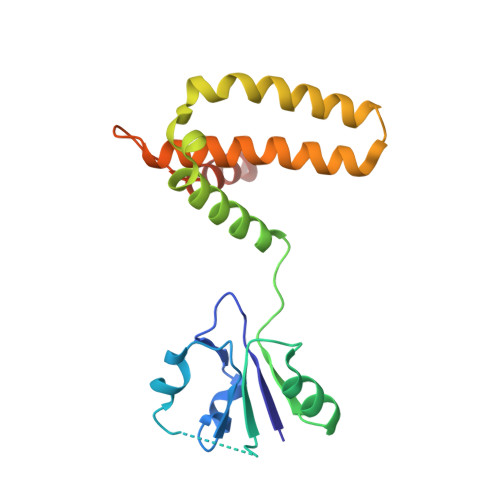
The housecleaning enzyme of Mycobacterium tuberculosis (Mtb), MazG, is a nucleoside triphosphate pyrophosphohydrolase (NTP-PPase) and can hydrolyze all canonical or non-canonical NTPs into NMPs and pyrophosphate. The Mycobacterium tuberculosis MazG (Mtb-MazG) contributes to antibiotic resistance in response to oxidative or nitrosative stress under dormancy, making it a promising target for treating TB in latent infection patients. However, the structural basis of Mtb-MazG is not clear. Here we describe the crystal structure of Mtb-MazG (1-185) at 2.7 Å resolution, composed of two similar folded spherical domains in tandem. Unlike other all-α NTP pyrophosphatases, Mtb-MazG has an N-terminal extra region composed of three α-helices and five β-strands. The second domain is global, with five α-helices located in the N-terminal domain. Gel-filtration assay and SAXS analysis show that Mtb-MazG forms an enzyme-active dimer in solution. In addition, the metal ion Mg 2+ is bound with four negative-charged residues Glu119, Glu122, Glu138, and Asp141. Different truncations and site-directed mutagenesis revealed that the full-length dimeric form and the metal ion Mg 2+ are indispensable for the catalytic activity of Mtb-MazG. Thus, our work provides new insights into understanding the molecular basis of Mtb - MazG.
Organizational Affiliation:
State Key Laboratory of Genetic Engineering, School of Life Sciences and Huashan Hospital, MOE Engineering Research Center of Gene Technology, Shanghai Engineering Research Center of Industrial Microorganisms, Fudan University, Shanghai, China.