Secreted in a Type III Secretion System-Dependent Manner, EsaH and EscE Are the Cochaperones of the T3SS Needle Protein EsaG of Edwardsiella piscicida.
Zeng, Z.X., Liu, L.Y., Xiao, S.B., Lu, J.F., Liu, Y.L., Li, J., Zhou, Y.Z., Liao, L.J., Li, D.Y., Zhou, Y., Nie, P., Xie, H.X.(2022) mBio 13: e0125022-e0125022
- PubMed: 35861543
- DOI: https://doi.org/10.1128/mbio.01250-22
- Primary Citation of Related Structures:
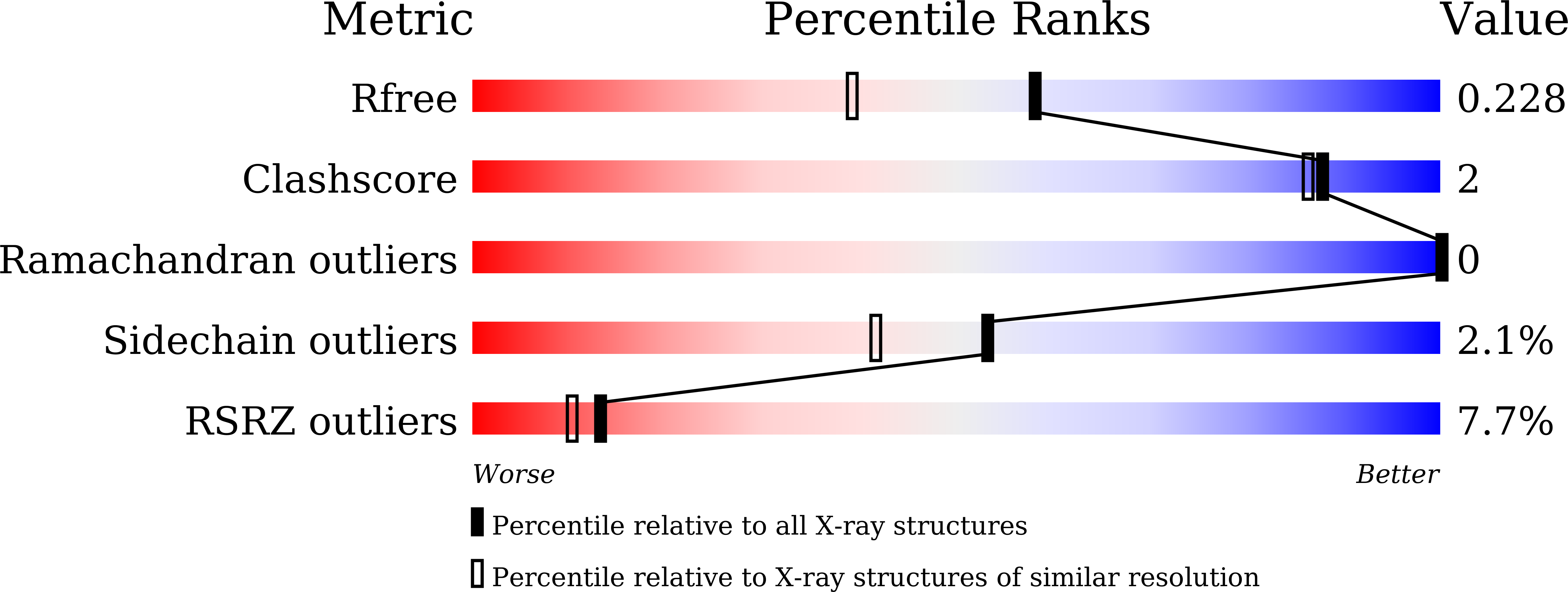
7Y6B, 7Y6C - PubMed Abstract:
The intracellular EscE protein tightly controls the secretion of the type III secretion system (T3SS) middle and late substrates in Edwardsiella piscicida. However, the regulation of secretion by EscE is incompletely understood. In this work, we reveal that EscE interacts with EsaH and EsaG. The crystal structures of the EscE-EsaH complex and EscE-EsaG-EsaH complex were resolved at resolutions of 1.4 Å and 1.8 Å, respectively. EscE and EsaH form a hydrophobic groove to engulf the C-terminal region of EsaG (56 to 73 amino acids [aa]), serving as the cochaperones of T3SS needle protein EsaG in E. piscicida . V61, K62, M64, and M65 of EsaG play a pivotal role in maintaining the conformation of the ternary complex of EscE-EsaG-EsaH, thereby maintaining the stability of EsaG. An in vivo experiment revealed that EscE and EsaH stabilize each other, and both of them stabilize EsaG. Meanwhile, either EscE or EsaH can be secreted through the T3SS. The secondary structure of EsaH lacks the fourth and fifth α helices presented in its homologs PscG, YscG, and AscG. Insertion of the α4 and α5 helices of PscG or swapping the N-terminal 25 aa of PscG with those of EsaH starkly decreases the protein level of the chimeric EsaH, resulting in instability of EsaG and deactivation of the T3SS. To the best of our knowledge, these data represent the first reported structure of the T3SS needle complex of pathogens from Enterobacteriaceae and the first evidence for the secretion of T3SS needle chaperones. IMPORTANCE Edwardsiella piscicida causes severe hemorrhagic septicemia in fish. Inactivation of the type III secretion system (T3SS) increases its 50% lethal dose (LD 50 ) by ~10 times. The secretion of T3SS middle and late substrates in E. piscicida is tightly controlled by the intracellular steady-state protein level of EscE, but the mechanism is incompletely understood. In this study, EscE was found to interact with and stabilize EsaH in E. piscicida . The EscE-EsaH complex is structurally analogous to T3SS needle chaperones. Further study revealed that EscE and EsaH form a hydrophobic groove to engulf the C-terminal region of EsaG, serving as the cochaperones stabilizing the T3SS needle protein EsaG. Interestingly, both EscE and EsaH are secreted. Our study reveals that the EscE-EsaH complex controls T3SS protein secretion by stabilizing EsaG, whose secretion in turn leads to the secretion of the middle and late T3SS substrates.
Organizational Affiliation:
College of Bioengineering, Qilu University of Technology, Jinan, Shandong Province, China.