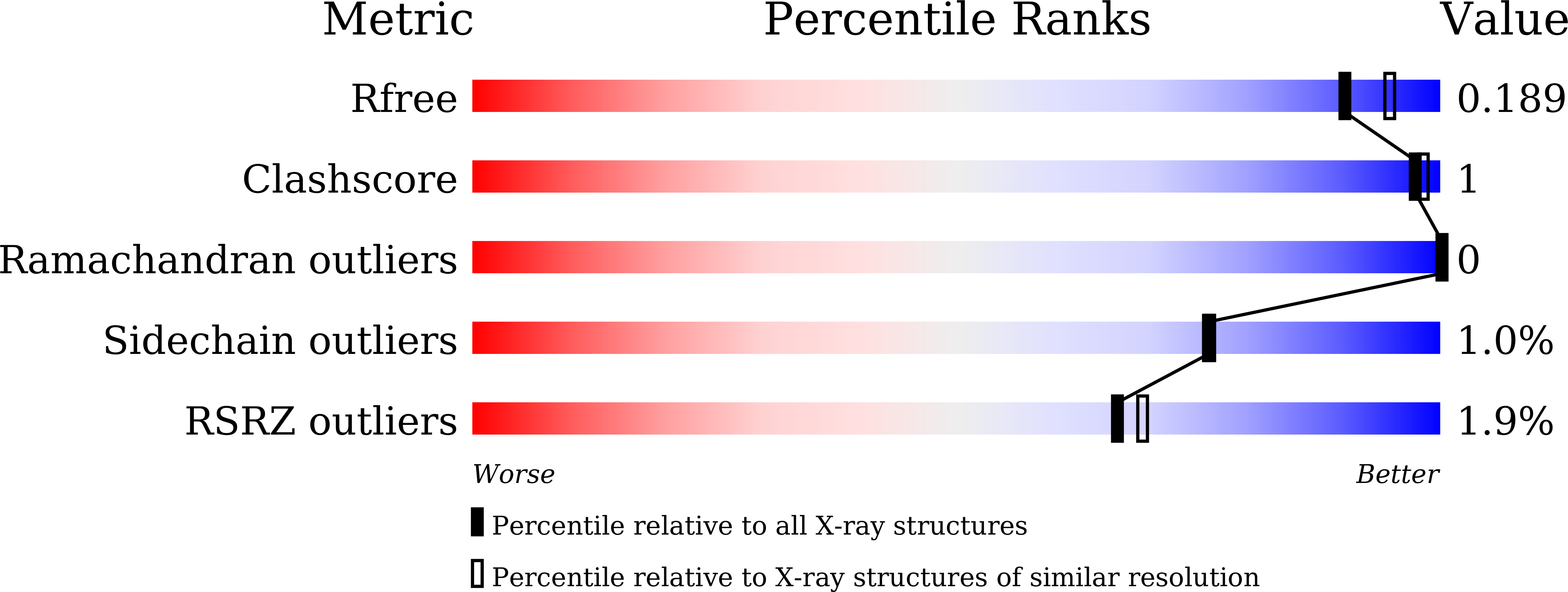
Discovery of the Biosynthetic Pathway of Beticolin 1 Reveals a Novel Non-Heme Iron-Dependent Oxygenase for Anthraquinone Ring Cleavage.
Hou, X., Xu, H., Deng, Z., Yan, Y., Yuan, Z., Liu, X., Su, Z., Yang, S., Zhang, Y., Rao, Y.(2022) Angew Chem Int Ed Engl 61: e202208772-e202208772
- PubMed: 35862137
- DOI: https://doi.org/10.1002/anie.202208772
- Primary Citation of Related Structures:
7Y3W, 7Y3X, 7Y3Y - PubMed Abstract:
This study used light-mediated comparative transcriptomics to identify the biosynthetic gene cluster of beticolin 1 in Cercospora. It contains an anthraquinone moiety and an unusual halogenated xanthone moiety connected by a bicyclo[3.2.2]nonane. During elucidation of the biosynthetic pathway of beticolin 1, a novel non-heme iron oxygenase BTG13 responsible for anthraquinone ring cleavage was discovered. More importantly, the discovery of non-heme iron oxygenase BTG13 is well supported by experimental evidence: (i) crystal structure and the inductively coupled plasma mass spectrometry revealed that its reactive site is built by an atypical iron ion coordination, where the iron ion is uncommonly coordinated by four histidine residues, an unusual carboxylated-lysine (Kcx377) and water; (ii) Kcx377 is mediated by His58 and Thr299 to modulate the catalytic activity of BTG13. Therefore, we believed this study updates our knowledge of metalloenzymes.
Organizational Affiliation:
Key Laboratory of Carbohydrate Chemistry and Biotechnology, Ministry of Education, School of Biotechnology, Jiangnan University, Wuxi, 214122, P. R. China.