Structural insights into the redox regulation of Oncomelania hupensis TRP14 and its potential role in the snail host response to parasite invasion.
Huang, S., Wang, S., Su, Z., Cao, Y., Hong, W., Lin, T.(2022) Fish Shellfish Immunol 128: 474-483
- PubMed: 35988710
- DOI: https://doi.org/10.1016/j.fsi.2022.08.040
- Primary Citation of Related Structures:
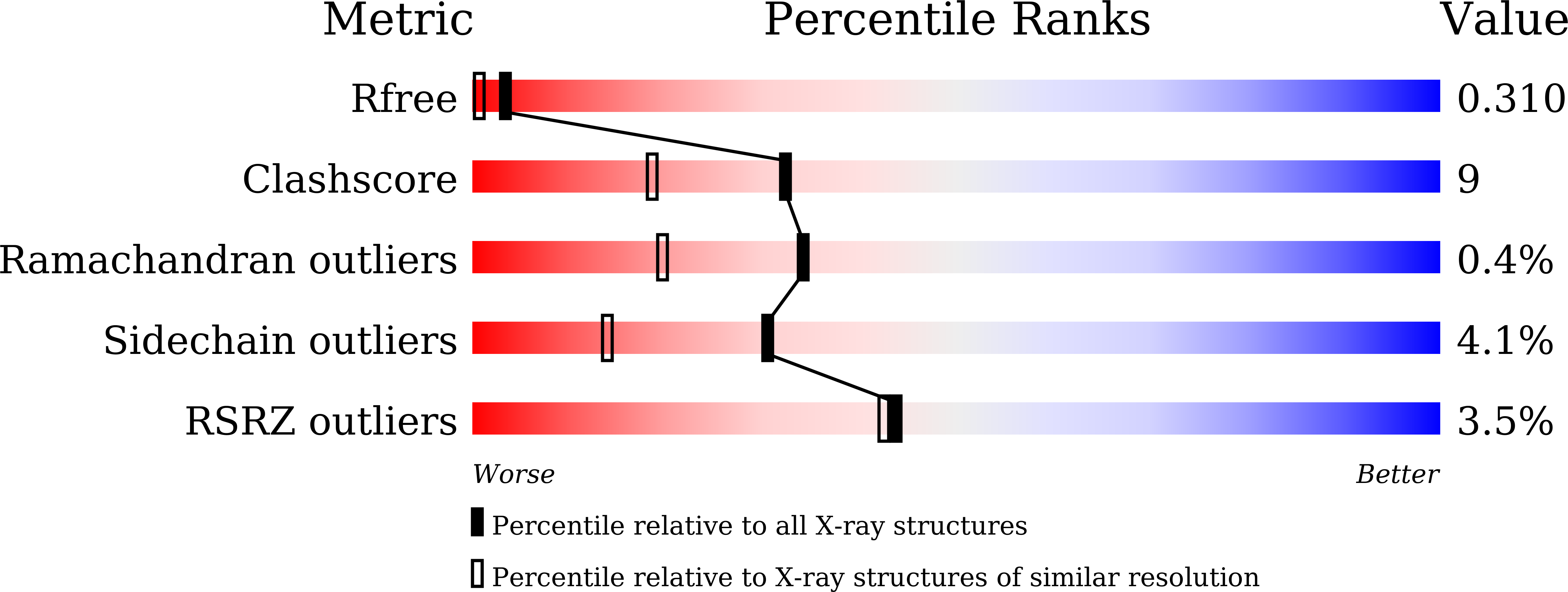
7XPW, 7XQ3 - PubMed Abstract:
The freshwater amphibious snail Oncomelania hupensis is the unique intermediate host of Schistosoma japonicum, but little attention has been paid to the interaction between the two. In snails, the production of reactive oxygen species (ROS) by hemocytes has been shown to be vital for snail immune defense against schistosome infection. However, excessive ROS accumulation could lead to oxidative damage, requiring the antioxidant system for maintaining the cellular redox homeostasis. Previously we identified a thioredoxin-related protein of 14 kDa from O. hupensis (OhTRP14), and showed that it was involved in the scavenging of ROS in circulating hemocytes. Here, we confirmed that OhTRP14 plays a potential role in the snail host response to parasite challenge and determined the crystal structures of OhTRP14 in two different states (oxidized and transition state). The overall structure revealed a typical Trx fold and is similar to that of human TRP14 (hTRP14), but there were significant structural differences between the two states. Noticeably, there was a different pair of thiol groups from Cys30 and Cys44 in the transition state of OhTRP14, were with the similar separation of 2.9 Å as that (2.6 Å) between Cys41 and Cys44, but in a different orientation, suggesting that the Cys30 is likely to function as an important molecular switch involved in the oxidoreductase activity of OhTRP14. Comparative studies between OhTRP14 and hTRP14 by analyzing the surface characteristics, charge distribution and oxidoreductase activity toward insulin demonstrated they might have similar substrates. The results are expected to provide structural insights into the redox regulation of OhTRP14 and contribute to better understanding of TRP14 family. DATA DEPOSITION: The atomic coordinates of the structure and the structure factors were deposited in Protein Data Bank with PDB ID codes 7XQ3 and 7XPW.
Organizational Affiliation:
Department of Parasitology, School of Basic Medical Sciences, Central South University, Changsha, Hunan, China. Electronic address: huangshuaiqin@xmu.edu.cn.