Structural insights into the interactions between lloviu virus VP30 and nucleoprotein.
Sun, W., Luan, F., Wang, J., Ma, L., Li, X., Yang, G., Hao, C., Qin, X., Dong, S.(2022) Biochem Biophys Res Commun 616: 82-88
- PubMed: 35649303
- DOI: https://doi.org/10.1016/j.bbrc.2022.05.059
- Primary Citation of Related Structures:
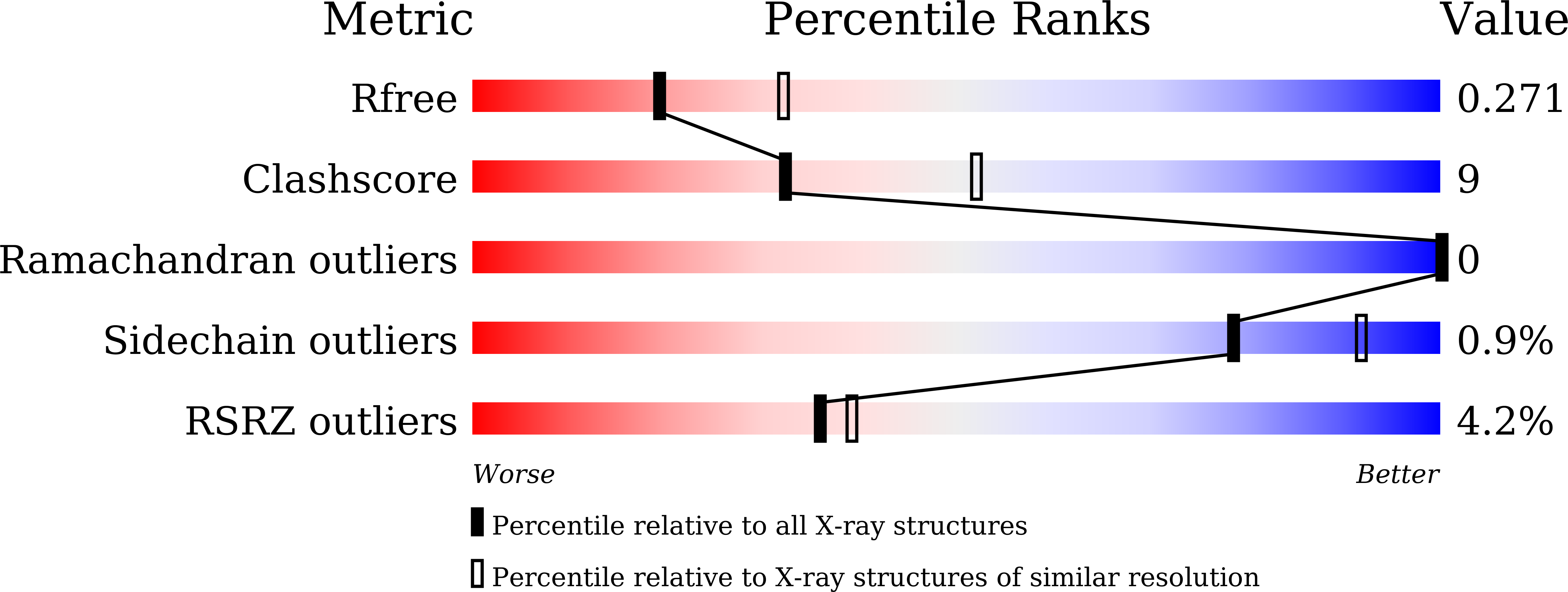
7XIV - PubMed Abstract:
The family Filoviridae comprises many notorious viruses, such as Ebola virus (EBOV) and Marburg virus (MARV), that can infect humans and nonhuman primates. Lloviu virus (LLOV), a less well studied filovirus, is considered a potential pathogen for humans. The VP30 C-terminal domain (CTD) of these filoviruses exhibits nucleoprotein (NP) binding and plays an essential role in viral transcription, replication and assembly. In this study, we confirmed the interactions between LLOV VP30 CTD and its NP fragment, and also determined the crystal structure of the chimeric dimeric LLOV NP-VP30 CTD at 2.50 Å resolution. The structure is highly conserved across the family Filoviridae. While in the dimer structure, only one VP30 CTD binds the NP fragment, which indicates that the interaction between LLOV VP30 CTD and NP is not strong. Our work provides a preliminary model to investigate the interactions between LLOV VP30 and NP and suggests a potential target for anti-filovirus drug development.
Organizational Affiliation:
School of Biological Science and Technology, University of Jinan, Jinan, China.