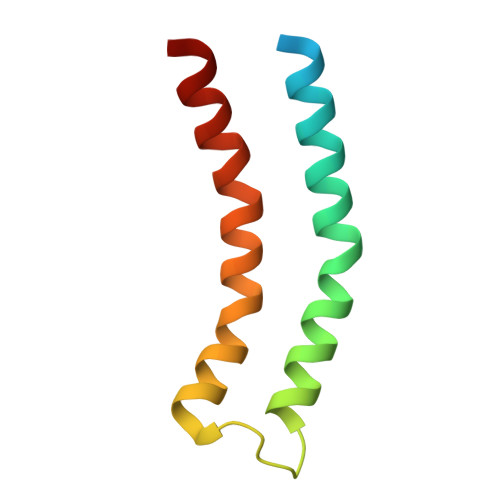
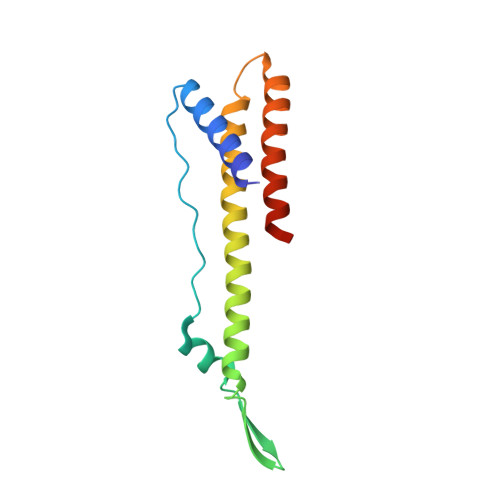
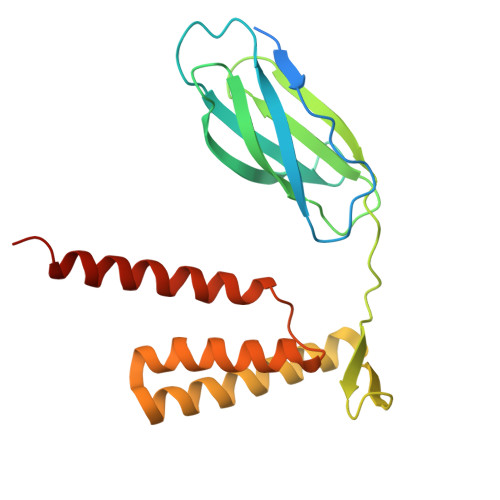
Structural insights into a spindle-shaped archaeal virus with a sevenfold symmetrical tail.
Han, Z., Yuan, W., Xiao, H., Wang, L., Zhang, J., Peng, Y., Cheng, L., Liu, H., Huang, L.(2022) Proc Natl Acad Sci U S A 119: e2119439119-e2119439119
- PubMed: 35895681
- DOI: https://doi.org/10.1073/pnas.2119439119
- Primary Citation of Related Structures:
7XDI - PubMed Abstract:
Archaeal viruses with a spindle-shaped virion are abundant and widespread in extremely diverse environments. However, efforts to obtain the high-resolution structure of a spindle-shaped virus have been unsuccessful. Here, we present the structure of SSV19, a spindle-shaped virus infecting the hyperthermophilic archaeon Sulfolobus sp. E11-6. Our near-atomic structure reveals an unusual sevenfold symmetrical virus tail consisting of the tailspike, nozzle, and adaptor proteins. The spindle-shaped capsid shell is formed by seven left-handed helical strands, constructed of the hydrophobic major capsid protein, emanating from the highly glycosylated tail assembly. Sliding between adjacent strands is responsible for the variation of a virion in size. Ultrathin sections of the SSV19-infected cells show that SSV19 virions adsorb to the host cell membrane through the tail after penetrating the S-layer. The tailspike harbors a putative endo-mannanase domain, which shares structural similarity to a Bacteroides thetaiotaomicro endo-mannanase. Molecules of glycerol dibiphytanyl glycerol tetraether lipid were observed in hydrophobic clefts between the tail and the capsid shell. The nozzle protein resembles the stem and clip domains of the portals of herpesviruses and bacteriophages, implying an evolutionary relationship among the archaeal, bacterial, and eukaryotic viruses.
Organizational Affiliation:
Key Laboratory for Matter Microstructure and Function of Hunan Province, Institute of Interdisciplinary Studies, Key Laboratory of Low-dimensional Quantum Structures and Quantum Control, School of Physics and Electronics, Hunan Normal University, Changsha, 410081 China.