Dual roles and evolutionary implications of P26/poxin in antagonizing intracellular cGAS-STING and extracellular melanization immunity.
Yin, M., Kuang, W., Wang, Q., Wang, X., Yuan, C., Lin, Z., Zhang, H., Deng, F., Jiang, H., Gong, P., Zou, Z., Hu, Z., Wang, M.(2022) Nat Commun 13: 6934-6934
- PubMed: 36376305
- DOI: https://doi.org/10.1038/s41467-022-34761-0
- Primary Citation of Related Structures:
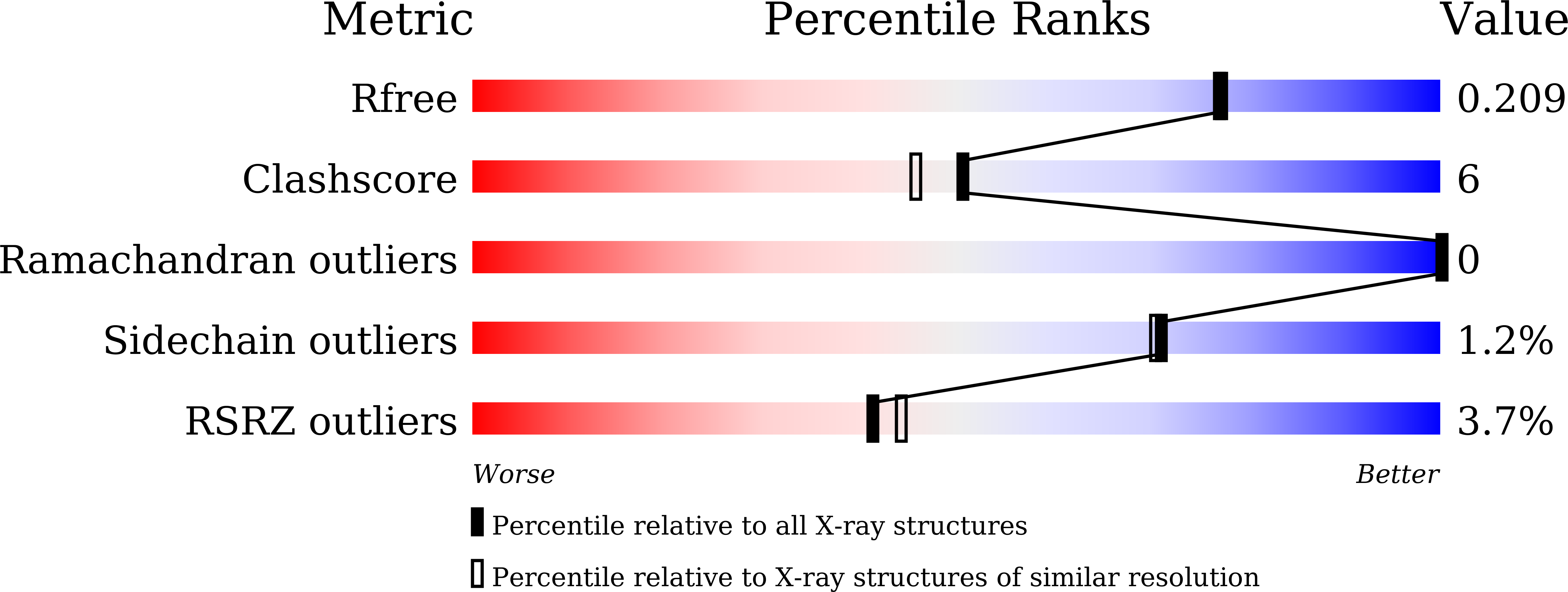
7WN7 - PubMed Abstract:
P26, a homolog of the viral-encoded nuclease poxin that neutralizes the cGAS-STING innate immunity, is widely distributed in various invertebrate viruses, lepidopteran insects, and parasitoid wasps. P26/poxin from certain insect viruses also retains protease activity, though its biological role remains unknown. Given that many P26s contain a signal peptide, it is surmised that P26 may possess certain extracellular functions. Here, we report that a secretory baculoviral P26 suppresses melanization, a prominent insect innate immunity against pathogen invasion. P26 targets the cofactor of a prophenoloxidase-activating protease, and its inhibitory function is independent of nuclease activity. The analysis of P26/poxin homologs from different origins suggests that the ability to inhibit the extracellular melanization pathway is limited to P26s with a signal peptide and not shared by the homologs without it. These findings highlight the independent evolution of a single viral suppressor to perform dual roles in modulating immunity during virus-host adaptation.
Organizational Affiliation:
State Key Laboratory of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China.