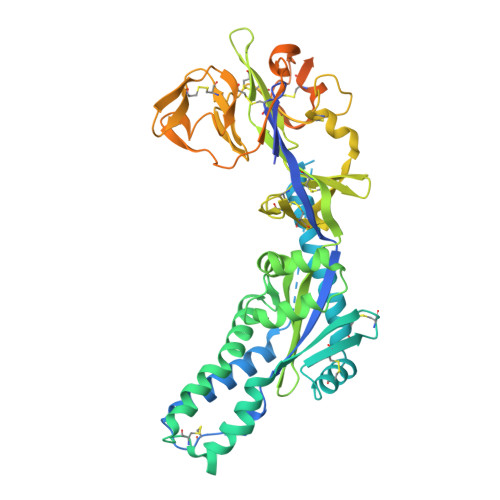
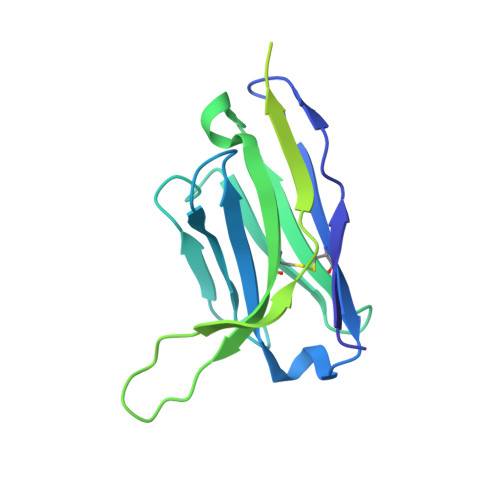
Characterization of prefusion-F-specific antibodies elicited by natural infection with human metapneumovirus.
Rush, S.A., Brar, G., Hsieh, C.L., Chautard, E., Rainho-Tomko, J.N., Slade, C.D., Bricault, C.A., Kume, A., Kearns, J., Groppo, R., Mundle, S.T., Zhang, L., Casimiro, D., Fu, T.M., DiNapoli, J.M., McLellan, J.S.(2022) Cell Rep 40: 111399-111399
- PubMed: 36130517
- DOI: https://doi.org/10.1016/j.celrep.2022.111399
- Primary Citation of Related Structures:
7TJQ, 7TL0 - PubMed Abstract:
Human metapneumovirus (hMPV) is a major cause of acute respiratory infections in infants and older adults, for which no vaccines or therapeutics are available. The viral fusion (F) glycoprotein is required for entry and is the primary target of neutralizing antibodies; however, little is known about the humoral immune response generated from natural infection. Here, using prefusion-stabilized F proteins to interrogate memory B cells from two older adults, we obtain over 700 paired non-IgM antibody sequences representing 563 clonotypes, indicative of a highly polyclonal response. Characterization of 136 monoclonal antibodies reveals broad recognition of the protein surface, with potently neutralizing antibodies targeting each antigenic site. Cryo-EM studies further reveal two non-canonical sites and the molecular basis for recognition of the apex of hMPV F by two prefusion-specific neutralizing antibodies. Collectively, these results provide insight into the humoral response to hMPV infection in older adults and will help guide vaccine development.
Organizational Affiliation:
Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX 78712, USA.