Crystal structure of a putative 3-hydroxypimelyl-CoA dehydrogenase, Hcd1, from Syntrophus aciditrophicus strain SB at 1.78 A resolution
Dinh, D.M., Thomas, L.M., Karr, E.A.(2023) Acta Crystallogr F Struct Biol Commun 79: 151-158
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2023) Acta Crystallogr F Struct Biol Commun 79: 151-158
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 3-oxoacyl-reductase | 267 | Syntrophus aciditrophicus SB | Mutation(s): 0 Gene Names: SYN_01680 EC: 1.1.1.100 | 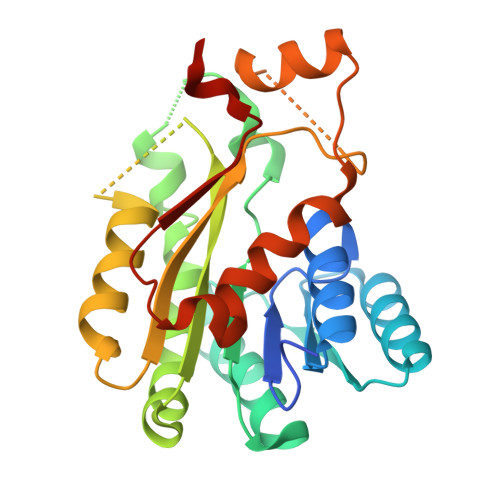 | |
UniProt | |||||
Find proteins for Q2LXS6 (Syntrophus aciditrophicus (strain SB)) Explore Q2LXS6 Go to UniProtKB: Q2LXS6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q2LXS6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GOL Query on GOL | B [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 56.006 | α = 90 |
| b = 56.006 | β = 90 |
| c = 133.129 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Department of Energy (DOE, United States) | United States | DE-FG02-96ER20214 |