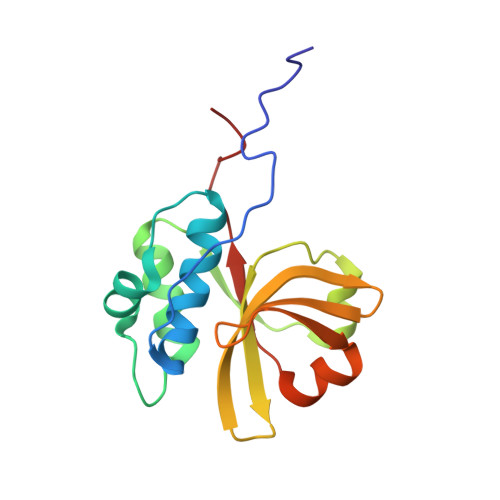
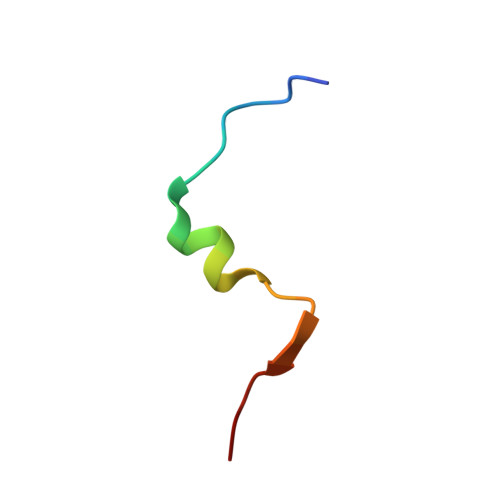
Structural and dynamic studies of the peptidase domain from Clostridium thermocellum PCAT1.
Bhattacharya, S., Palillo, A.(2022) Protein Sci 31: 498-512
- PubMed: 34865273
- DOI: https://doi.org/10.1002/pro.4248
- Primary Citation of Related Structures:
7N87, 7S5J - PubMed Abstract:
The export of antimicrobial peptides is mediated by diverse mechanisms in bacterial quorum sensing pathways. One such binary system employed by gram-positive bacteria is the PCAT1 ABC transporter coupled to a cysteine protease. The focus of this study is the N-terminal C39 peptidase (PEP) domain from Clostridium thermocellum PCAT1 that processes its natural substrate CtA by cleaving a conserved -GG- motif to separate the cargo from the leader peptide prior to secretion. In this study, we are primarily interested in elucidating the dynamic and structural determinants of CtA binding and how it is coupled to cleavage efficiency in the PCAT1 PEP domain. To this end, we have characterized CtA interactions with PEP domain and PCAT1 transporter in detergent micelles using solution nuclear magnetic resonance spectroscopy. The bound CtA structure revealed the disordered C-terminal cargo peptide is linked by a sterically hindered cleavage site to a helix docked within a hydrophobic cavity in the PEP domain. The wide range of internal motions detected by amide nitrogen (N 15 ) relaxation measurements in the free enzyme and substrate-bound complex suggests the binding site is relatively floppy. This flexibility plays a key role in the structural rearrangement necessary to relax steric inhibition in the bound substrate. In conjunction with previously reported PCAT1 structures, we offer fresh insight into the ATP-mediated association between PEP and transmembrane domains as a putative mechanism to optimize peptide cleavage by regulating the width and flexibility of the enzyme active site.
Organizational Affiliation:
New York Structural Biology Center, New York, New York, USA.