Novel fold of rotavirus glycan-binding domain predicted by AlphaFold2 and determined by X-ray crystallography.
Hu, L., Salmen, W., Sankaran, B., Lasanajak, Y., Smith, D.F., Crawford, S.E., Estes, M.K., Prasad, B.V.V.(2022) Commun Biol 5: 419-419
- PubMed: 35513489
- DOI: https://doi.org/10.1038/s42003-022-03357-1
- Primary Citation of Related Structures:
7RSW - PubMed Abstract:
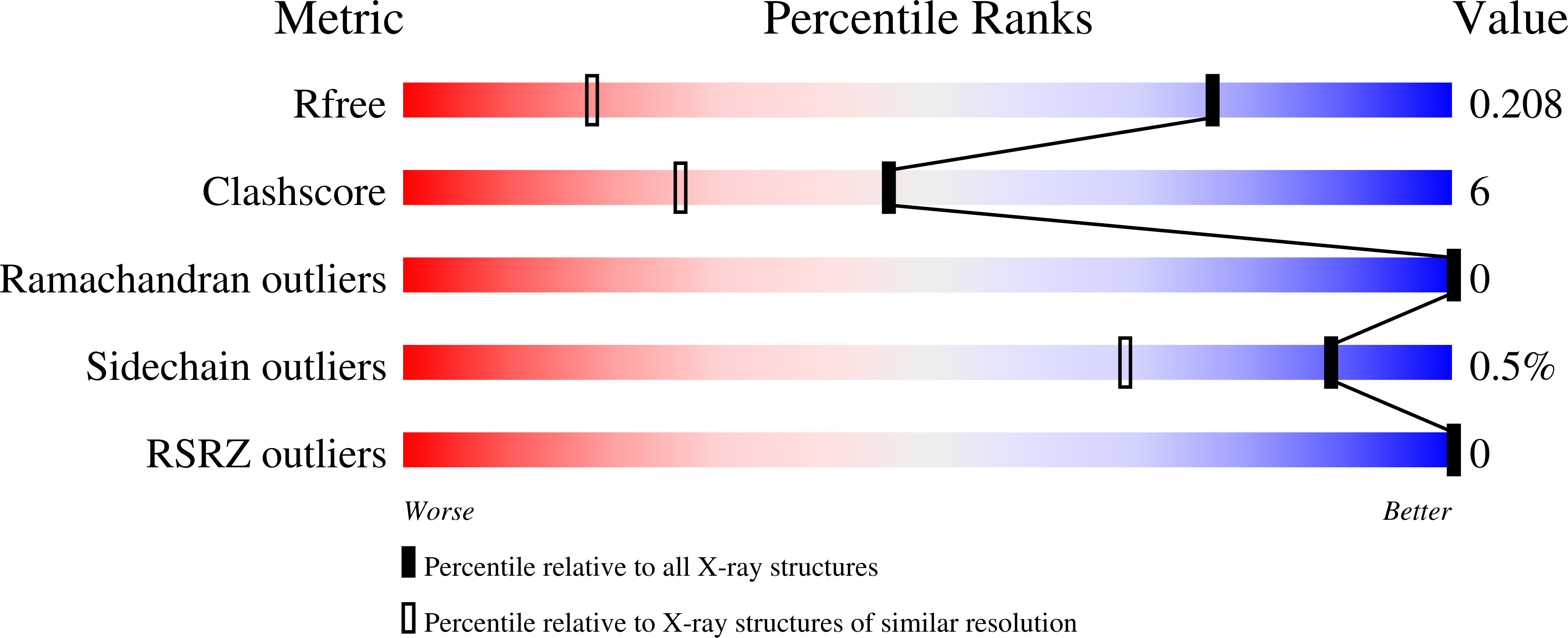
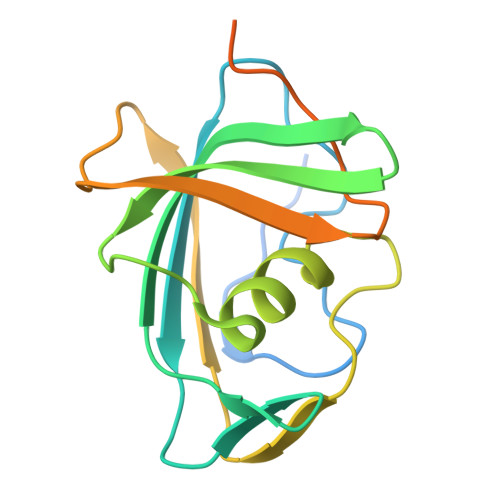
The VP8* domain of spike protein VP4 in group A and C rotaviruses, which cause epidemic gastroenteritis in children, exhibits a conserved galectin-like fold for recognizing glycans during cell entry. In group B rotavirus, which causes significant diarrheal outbreaks in adults, the VP8* domain (VP8*B) surprisingly lacks sequence similarity with VP8* of group A or group C rotavirus. Here, by using the recently developed AlphaFold2 for ab initio structure prediction and validating the predicted model by determining a 1.3-Å crystal structure, we show that VP8*B exhibits a novel fold distinct from the galectin fold. This fold with a β-sheet clasping an α-helix represents a new fold for glycan recognition based on glycan array screening, which shows that VP8*B recognizes glycans containing N-acetyllactosamine moiety. Although uncommon, our study illustrates how evolution can incorporate structurally distinct folds with similar functionality in a homologous protein within the same virus genus.
Organizational Affiliation:
Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, TX, USA. lhu@bcm.edu.