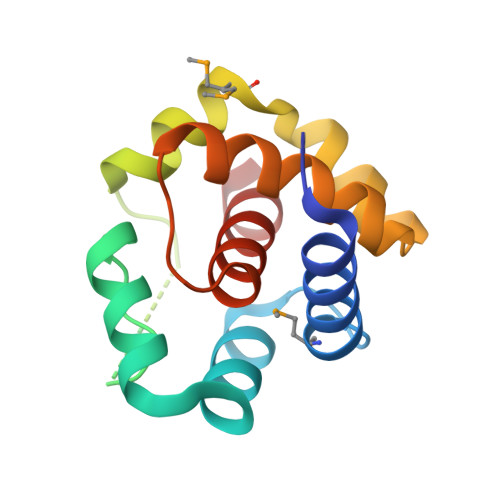
Structural and functional analysis of EntV reveals a 12 amino acid fragment protective against fungal infections.
Cruz, M.R., Cristy, S., Guha, S., De Cesare, G.B., Evdokimova, E., Sanchez, H., Borek, D., Miramon, P., Yano, J., Fidel Jr., P.L., Savchenko, A., Andes, D.R., Stogios, P.J., Lorenz, M.C., Garsin, D.A.(2022) Nat Commun 13: 6047-6047
- PubMed: 36229448
- DOI: https://doi.org/10.1038/s41467-022-33613-1
- Primary Citation of Related Structures:
7ROA - PubMed Abstract:
Fungal pathogens are a continuing challenge due to few effective antifungals and a rise in resistance. In previous work, we described the inhibition of Candida albicans virulence following exposure to the 68 amino acid bacteriocin, EntV, secreted by Enterococcus faecalis. Here, to optimize EntV as a potential therapeutic and better understand its antifungal features, an X-ray structure is obtained. The structure consists of six alpha helices enclosing a seventh 16 amino acid helix (α7). The individual helices are tested for antifungal activity using in vitro and nematode infection assays. Interestingly, α7 retains antifungal, but not antibacterial activity and is also effective against Candida auris and Cryptococcus neoformans. Further reduction of α7 to 12 amino acids retains full antifungal activity, and excellent efficacy is observed in rodent models of C. albicans oropharyngeal, systemic, and venous catheter infections. Together, these results showcase EntV-derived peptides as promising candidates for antifungal therapeutic development.
Organizational Affiliation:
Department of Microbiology and Molecular Genetics, The University of Texas Health Science Center at Houston, Houston, TX, 77030, USA.