The Bacillus subtilis open reading frame ysgA encodes the SPOUT methyltransferase RlmP forming 2'- O -methylguanosine at position 2553 in the A-loop of 23S rRNA.
Roovers, M., Labar, G., Wolff, P., Feller, A., Van Elder, D., Soin, R., Gueydan, C., Kruys, V., Droogmans, L.(2022) RNA 28: 1185-1196
- PubMed: 35710145
- DOI: https://doi.org/10.1261/rna.079131.122
- Primary Citation of Related Structures:
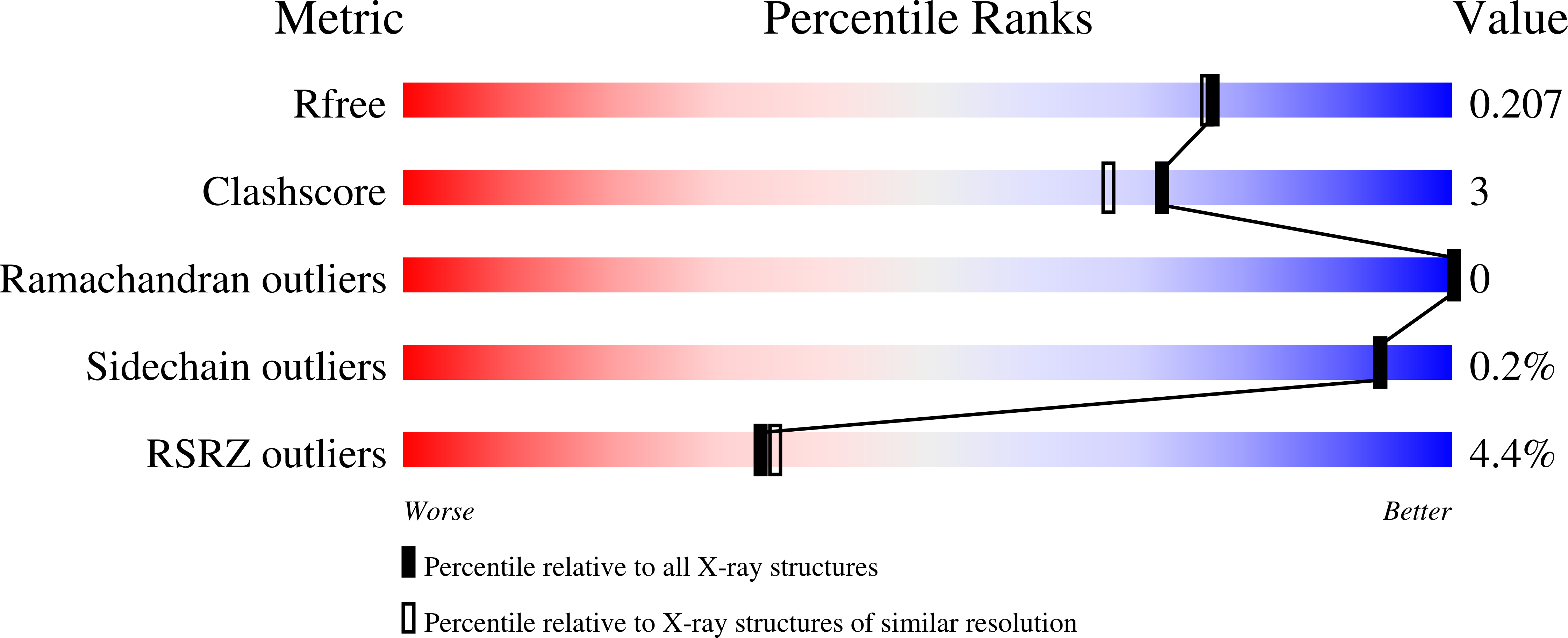
7QIU - PubMed Abstract:
A previous bioinformatic analysis predicted that the ysgA open reading frame of Bacillus subtilis encodes an RNA methyltransferase of the SPOUT superfamily. Here we show that YsgA is the 2'- O -methyltransferase that targets position G2553 ( Escherichia coli numbering) of the A-loop of 23S rRNA. This was shown by a combination of biochemical and mass spectrometry approaches using both rRNA extracted from B. subtilis wild-type or ΔysgA cells and in vitro synthesized rRNA. When the target G2553 is mutated, YsgA is able to methylate the ribose of adenosine. However, it cannot methylate cytidine nor uridine. The enzyme modifies free 23S rRNA but not the fully assembled ribosome nor the 50S subunit, suggesting that the modification occurs early during ribosome biogenesis. Nevertheless, ribosome subunits assembly is unaffected in a B. subtilis ΔysgA mutant strain. The crystal structure of the recombinant YsgA protein, combined with mutagenesis data, outlined in this article highlights a typical SPOUT fold preceded by an L7Ae/L30 (eL8/eL30 in a new nomenclature) amino-terminal domain.
Organizational Affiliation:
Labiris, B-1070 Bruxelles, Belgium.