A blast fungus zinc-finger fold effector binds to a hydrophobic pocket in host Exo70 proteins to modulate immune recognition in rice.
De la Concepcion, J.C., Fujisaki, K., Bentham, A.R., Cruz Mireles, N., Sanchez de Medina Hernandez, V., Shimizu, M., Lawson, D.M., Kamoun, S., Terauchi, R., Banfield, M.J.(2022) Proc Natl Acad Sci U S A 119: e2210559119-e2210559119
- PubMed: 36252011
- DOI: https://doi.org/10.1073/pnas.2210559119
- Primary Citation of Related Structures:
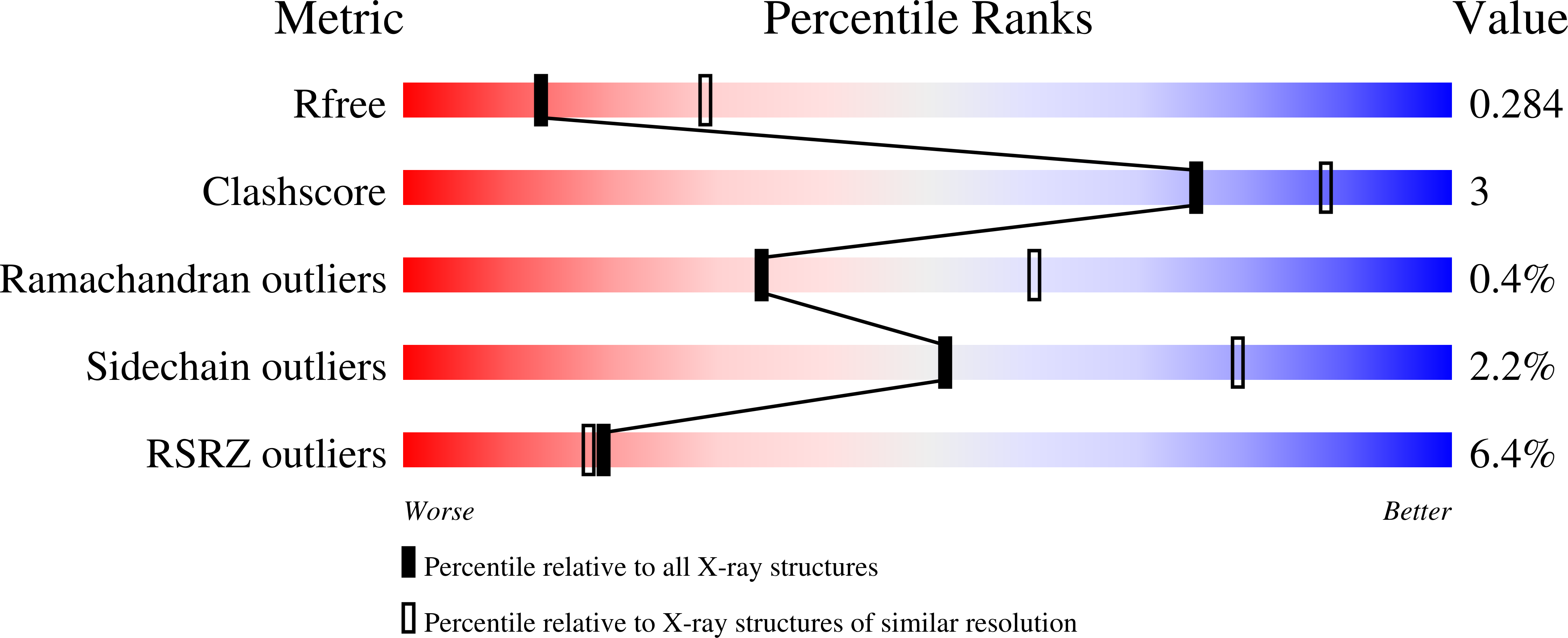
7PP2 - PubMed Abstract:
Exocytosis plays an important role in plant-microbe interactions, in both pathogenesis and symbiosis. Exo70 proteins are integral components of the exocyst, an octameric complex that mediates tethering of vesicles to membranes in eukaryotes. Although plant Exo70s are known to be targeted by pathogen effectors, the underpinning molecular mechanisms and the impact of this interaction on infection are poorly understood. Here, we show the molecular basis of the association between the effector AVR-Pii of the blast fungus Maganaporthe oryzae and rice Exo70 alleles OsExo70F2 and OsExo70F3, which is sensed by the immune receptor pair Pii via an integrated RIN4/NOI domain. The crystal structure of AVR-Pii in complex with OsExo70F2 reveals that the effector binds to a conserved hydrophobic pocket in Exo70, defining an effector/target binding interface. Structure-guided and random mutagenesis validates the importance of AVR-Pii residues at the Exo70 binding interface to sustain protein association and disease resistance in rice when challenged with fungal strains expressing effector mutants. Furthermore, the structure of AVR-Pii defines a zinc-finger effector fold (ZiF) distinct from the MAX (Magnaporthe Avrs and ToxB-like) fold previously described for a majority of characterized M . oryzae effectors. Our data suggest that blast fungus ZiF effectors bind a conserved Exo70 interface to manipulate plant exocytosis and that these effectors are also baited by plant immune receptors, pointing to new opportunities for engineering disease resistance.
Organizational Affiliation:
Department of Biochemistry and Metabolism, John Innes Centre, Norwich, NR4 7UH, United Kingdom.