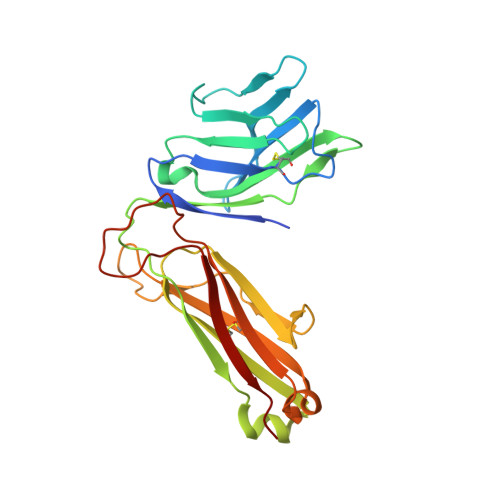
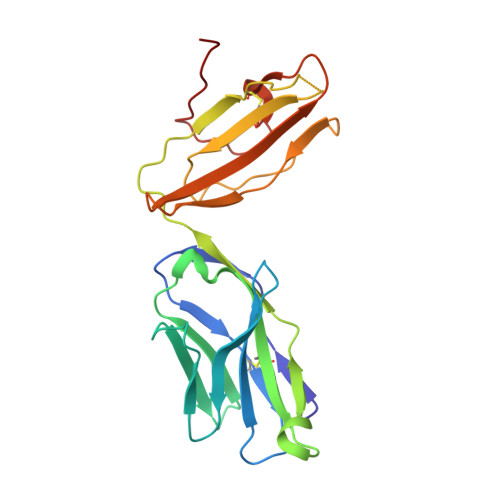
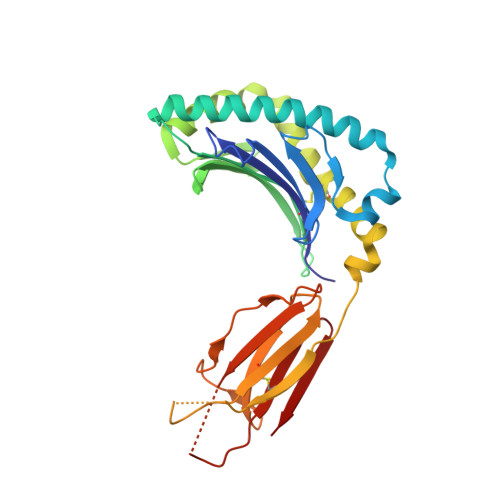
Recognition of the antigen-presenting molecule MR1 by a V delta 3 + gamma delta T cell receptor.
Rice, M.T., von Borstel, A., Chevour, P., Awad, W., Howson, L.J., Littler, D.R., Gherardin, N.A., Le Nours, J., Giles, E.M., Berry, R., Godfrey, D.I., Davey, M.S., Rossjohn, J., Gully, B.S.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34845016
- DOI: https://doi.org/10.1073/pnas.2110288118
- Primary Citation of Related Structures:
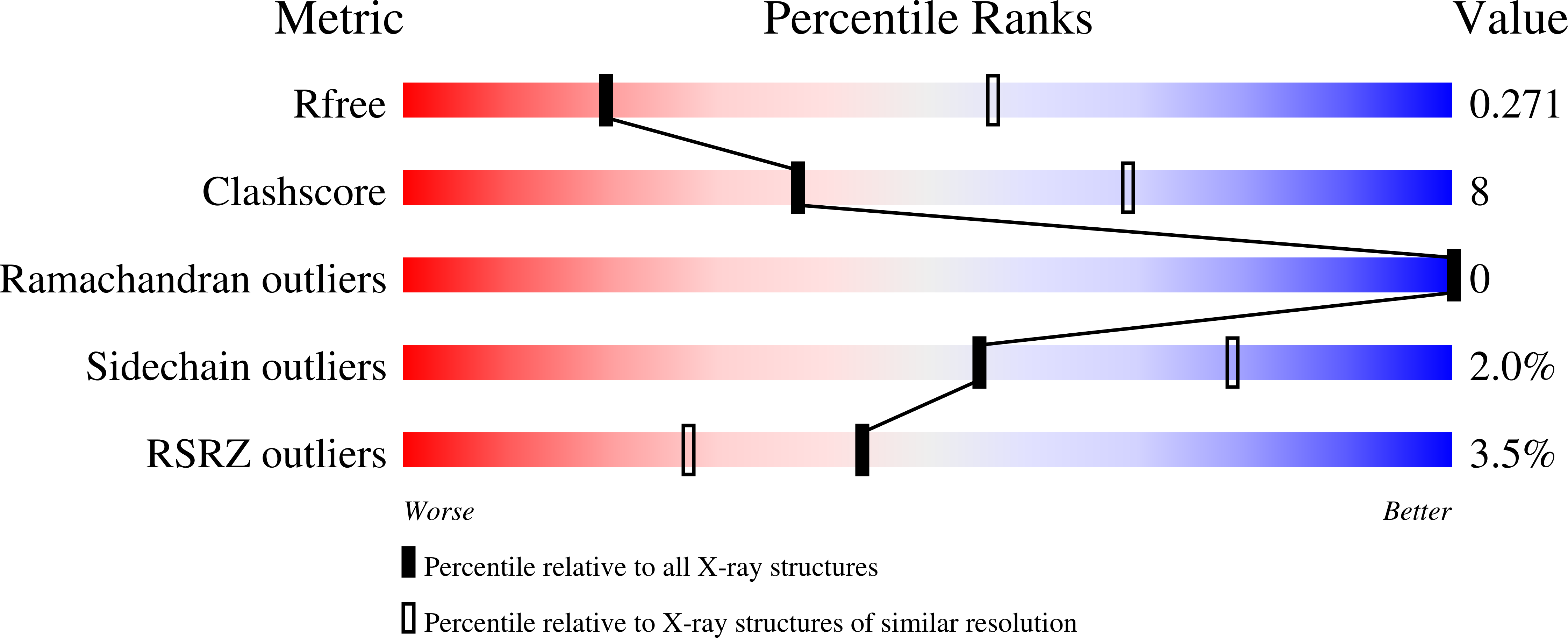
7LLI, 7LLJ - PubMed Abstract:
Unlike conventional αβ T cells, γδ T cells typically recognize nonpeptide ligands independently of major histocompatibility complex (MHC) restriction. Accordingly, the γδ T cell receptor (TCR) can potentially recognize a wide array of ligands; however, few ligands have been described to date. While there is a growing appreciation of the molecular bases underpinning variable (V)δ1 + and Vδ2 + γδ TCR-mediated ligand recognition, the mode of Vδ3 + TCR ligand engagement is unknown. MHC class I-related protein, MR1, presents vitamin B metabolites to αβ T cells known as mucosal-associated invariant T cells, diverse MR1-restricted T cells, and a subset of human γδ T cells. Here, we identify Vδ1/2 - γδ T cells in the blood and duodenal biopsy specimens of children that showed metabolite-independent binding of MR1 tetramers. Characterization of one Vδ3Vγ8 TCR clone showed MR1 reactivity was independent of the presented antigen. Determination of two Vδ3Vγ8 TCR-MR1-antigen complex structures revealed a recognition mechanism by the Vδ3 TCR chain that mediated specific contacts to the side of the MR1 antigen-binding groove, representing a previously uncharacterized MR1 docking topology. The binding of the Vδ3 + TCR to MR1 did not involve contacts with the presented antigen, providing a basis for understanding its inherent MR1 autoreactivity. We provide molecular insight into antigen-independent recognition of MR1 by a Vδ3 + γδ TCR that strengthens an emerging paradigm of antibody-like ligand engagement by γδ TCRs.
Organizational Affiliation:
Infection and Immunity Program, Biomedicine Discovery Institute, Monash University, Clayton, VIC 3800, Australia.