Functional and Structural Characterization of the UDP-Glucose Dehydrogenase Involved in Capsular Polysaccharide Biosynthesis from Campylobacter jejuni .
Riegert, A.S., Raushel, F.M.(2021) Biochemistry 60: 725-734
- PubMed: 33621065
- DOI: https://doi.org/10.1021/acs.biochem.0c00953
- Primary Citation of Related Structures:
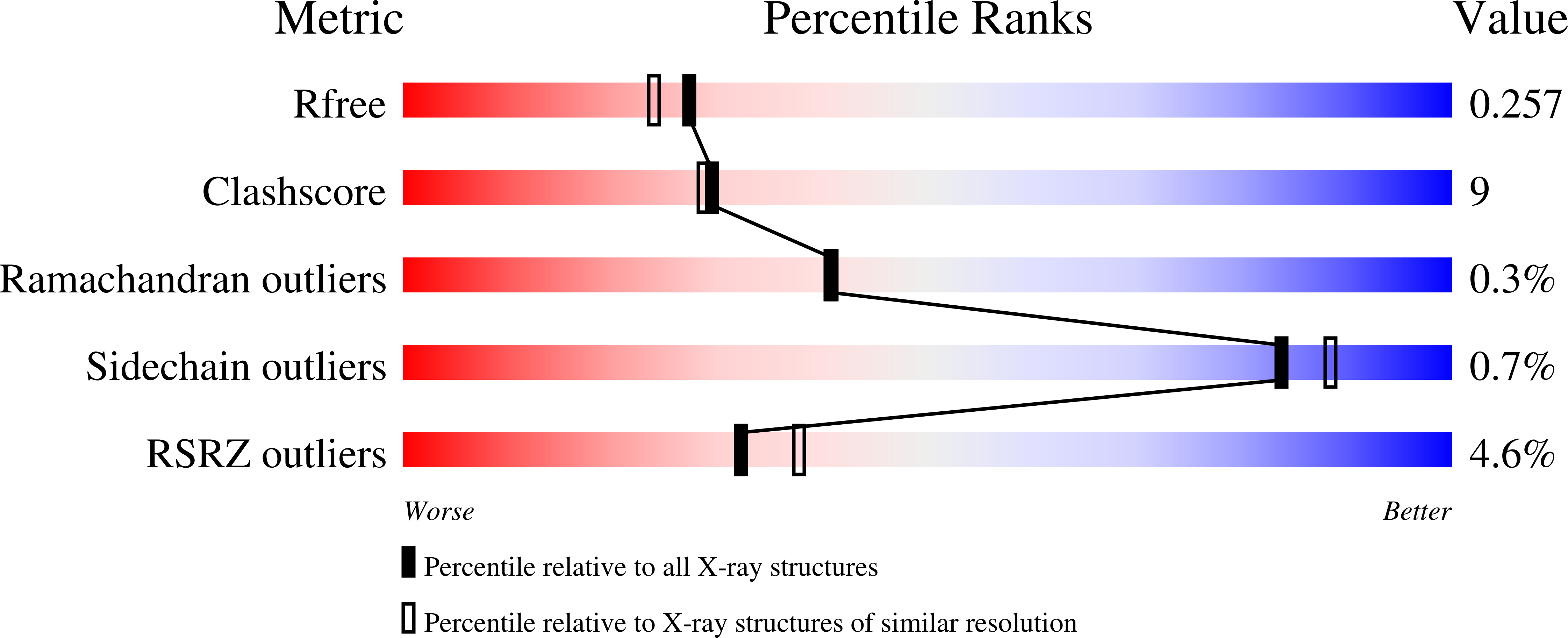
7KWS - PubMed Abstract:
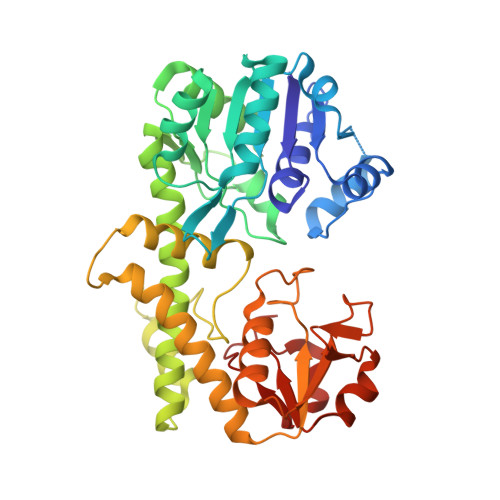
Campylobacter jejuni is a pathogenic organism that can cause campylobacteriosis in children and adults. Most commonly, campylobacter infection is brought on by consumption of raw or undercooked poultry, unsanitary drinking water, or pet feces. Surrounding the C. jejuni bacterium is a coat of sugar molecules known as the capsular polysaccharide (CPS). The capsular polysaccharide can be very diverse among the different strains of C. jejuni , and this diversity is considered important for evading the host immune system. Modifications to the CPS of C. jejuni NCTC 11168 include O-methylation, phosphoramidylation, and amidation of glucuronate with either serinol or ethanolamine. The enzymes responsible for amidation of glucuronate are currently unknown. In this study, Cj1441, an enzyme expressed from the CPS biosynthetic gene cluster in C. jejuni NCTC 11168, was shown to catalyze the oxidation of UDP-α-d-glucose into UDP-α-d-glucuronic acid with NAD + as the cofactor. No amide products were found in an attempt to determine whether the putative thioester intermediate formed during the oxidation of UDP-glucose by Cj1441 could be captured in the presence of added amines. The three-dimensional crystal structure of Cj1441 was determined in the presence of NAD + and UDP-glucose bound in the active site of the enzyme (Protein Data Bank entry 7KWS). A more thorough bioinformatic analysis of the CPS gene cluster suggests that the amidation activity is localized to the t-terminal half of Cj1438, a bifunctional enzyme that is currently annotated as a sugar transferase.
Organizational Affiliation:
Department of Biochemistry & Biophysics, Texas A&M University, College Station, Texas 77843, United States.