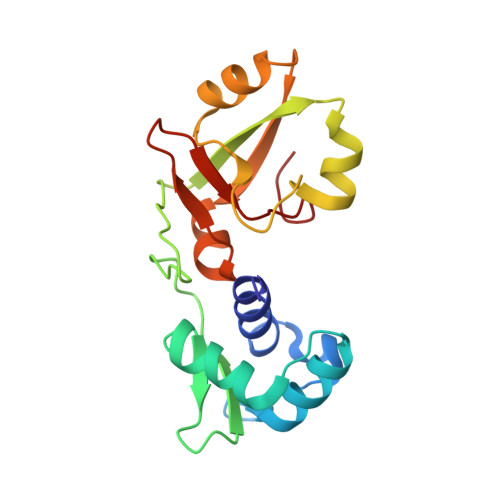
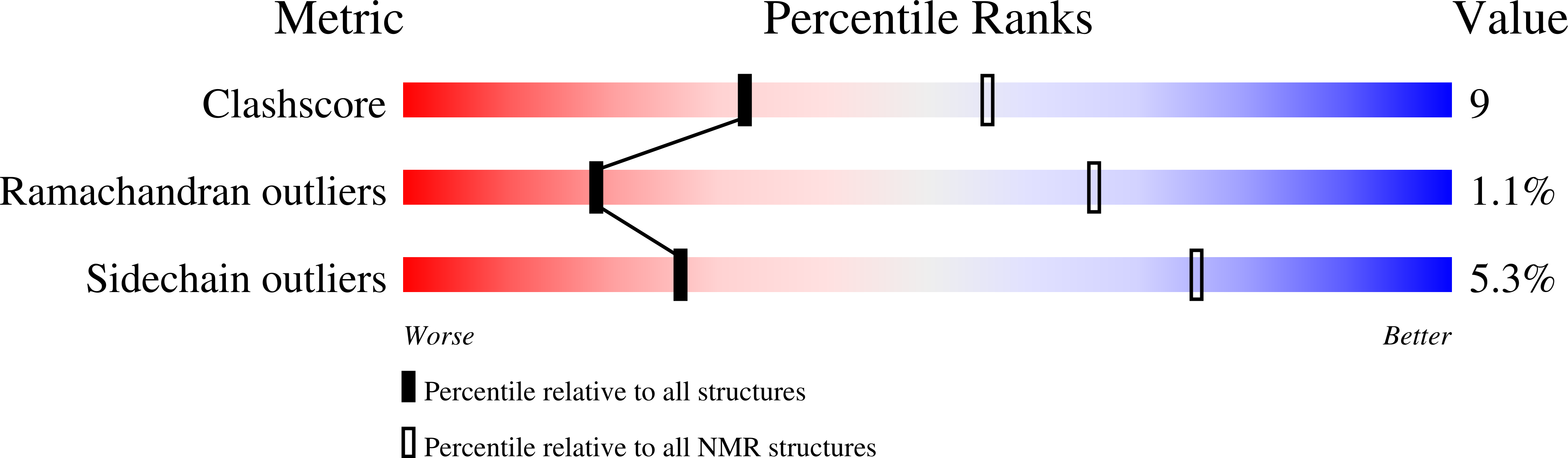NMR solution structures of Runella slithyformis RNA 2'-phosphotransferase Tpt1 provide insights into NAD+ binding and specificity.
Alphonse, S., Banerjee, A., Dantuluri, S., Shuman, S., Ghose, R.(2021) Nucleic Acids Res 49: 9607-9624
- PubMed: 33880546
- DOI: https://doi.org/10.1093/nar/gkab241
- Primary Citation of Related Structures:
7KW8, 7KW9 - PubMed Abstract:
Tpt1, an essential component of the fungal and plant tRNA splicing machinery, catalyzes transfer of an internal RNA 2'-PO4 to NAD+ yielding RNA 2'-OH and ADP-ribose-1',2'-cyclic phosphate products. Here, we report NMR structures of the Tpt1 ortholog from the bacterium Runella slithyformis (RslTpt1), as apoenzyme and bound to NAD+. RslTpt1 consists of N- and C-terminal lobes with substantial inter-lobe dynamics in the free and NAD+-bound states. ITC measurements of RslTpt1 binding to NAD+ (KD ∼31 μM), ADP-ribose (∼96 μM) and ADP (∼123 μM) indicate that substrate affinity is determined primarily by the ADP moiety; no binding of NMN or nicotinamide is observed by ITC. NAD+-induced chemical shift perturbations (CSPs) localize exclusively to the RslTpt1 C-lobe. NADP+, which contains an adenylate 2'-PO4 (mimicking the substrate RNA 2'-PO4), binds with lower affinity (KD ∼1 mM) and elicits only N-lobe CSPs. The RslTpt1·NAD+ binary complex reveals C-lobe contacts to adenosine ribose hydroxyls (His99, Thr101), the adenine nucleobase (Asn105, Asp112, Gly113, Met117) and the nicotinamide riboside (Ser125, Gln126, Asn163, Val165), several of which are essential for RslTpt1 activity in vivo. Proximity of the NAD+ β-phosphate to ribose-C1″ suggests that it may stabilize an oxocarbenium transition-state during the first step of the Tpt1-catalyzed reaction.
Organizational Affiliation:
Department of Chemistry and Biochemistry, The City College of New York, New York, NY 10031, USA.