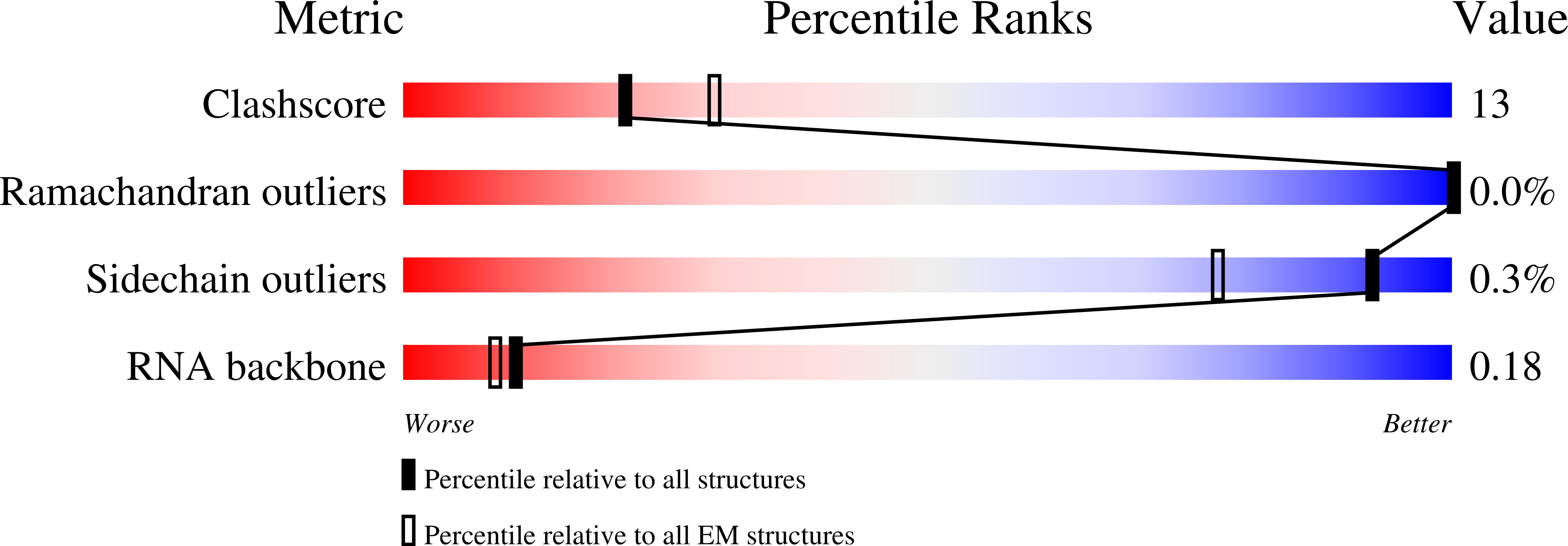
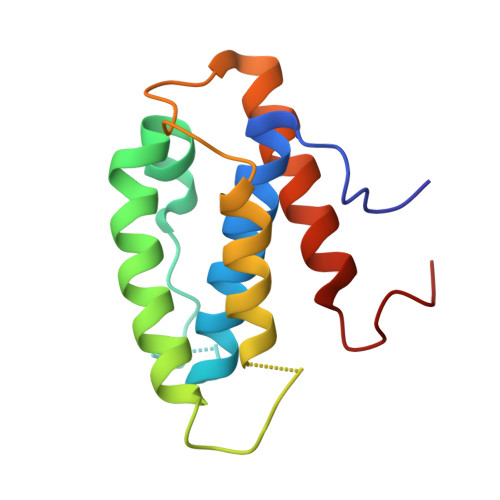
Structural basis for assembly of non-canonical small subunits into type I-C Cascade.
O'Brien, R.E., Santos, I.C., Wrapp, D., Bravo, J.P.K., Schwartz, E.A., Brodbelt, J.S., Taylor, D.W.(2020) Nat Commun 11: 5931-5931
- PubMed: 33230133
- DOI: https://doi.org/10.1038/s41467-020-19785-8
- Primary Citation of Related Structures:
7KHA - PubMed Abstract:
Bacteria and archaea employ CRISPR (clustered, regularly, interspaced, short palindromic repeats)-Cas (CRISPR-associated) systems as a type of adaptive immunity to target and degrade foreign nucleic acids. While a myriad of CRISPR-Cas systems have been identified to date, type I-C is one of the most commonly found subtypes in nature. Interestingly, the type I-C system employs a minimal Cascade effector complex, which encodes only three unique subunits in its operon. Here, we present a 3.1 Å resolution cryo-EM structure of the Desulfovibrio vulgaris type I-C Cascade, revealing the molecular mechanisms that underlie RNA-directed complex assembly. We demonstrate how this minimal Cascade utilizes previously overlooked, non-canonical small subunits to stabilize R-loop formation. Furthermore, we describe putative PAM and Cas3 binding sites. These findings provide the structural basis for harnessing the type I-C Cascade as a genome-engineering tool.
Organizational Affiliation:
Institute for Cell and Molecular Biology, University of Texas at Austin, Austin, TX, 78712, USA.