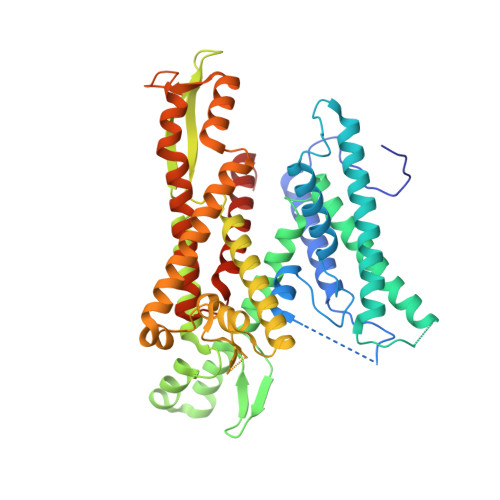
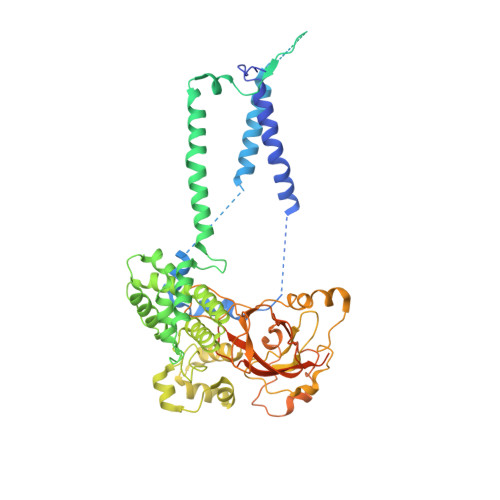
Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Itskanov, S., Kuo, K.M., Gumbart, J.C., Park, E.(2021) Nat Struct Mol Biol 28: 162-172
- PubMed: 33398175
- DOI: https://doi.org/10.1038/s41594-020-00541-x
- Primary Citation of Related Structures:
7KAH, 7KAI, 7KAJ, 7KAK, 7KAL, 7KAM, 7KAN, 7KAO, 7KAP, 7KAQ, 7KAR, 7KAS, 7KAT, 7KAU, 7KB5 - PubMed Abstract:
Many proteins are transported into the endoplasmic reticulum by the universally conserved Sec61 channel. Post-translational transport requires two additional proteins, Sec62 and Sec63, but their functions are poorly defined. In the present study, we determined cryo-electron microscopy (cryo-EM) structures of several variants of Sec61-Sec62-Sec63 complexes from Saccharomyces cerevisiae and Thermomyces lanuginosus and show that Sec62 and Sec63 induce opening of the Sec61 channel. Without Sec62, the translocation pore of Sec61 remains closed by the plug domain, rendering the channel inactive. We further show that the lateral gate of Sec61 must first be partially opened by interactions between Sec61 and Sec63 in cytosolic and luminal domains, a simultaneous disruption of which completely closes the channel. The structures and molecular dynamics simulations suggest that Sec62 may also prevent lipids from invading the channel through the open lateral gate. Our study shows how Sec63 and Sec62 work together in a hierarchical manner to activate Sec61 for post-translational protein translocation.
Organizational Affiliation:
Biophysics Graduate Program, University of California, Berkeley, CA, USA.