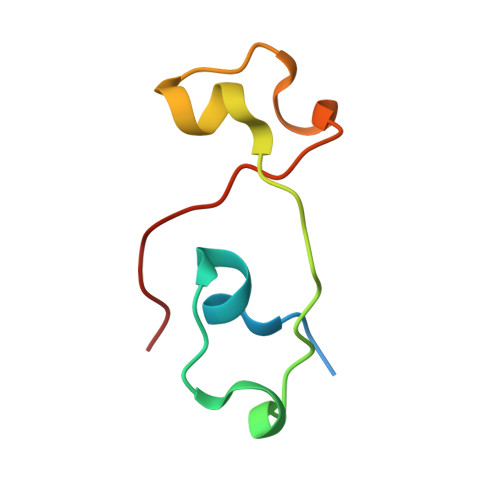
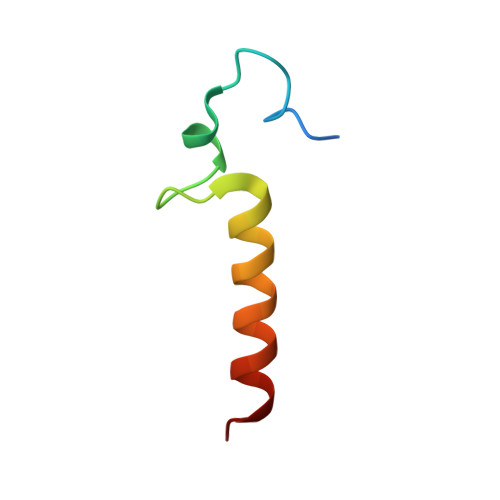
Molecular mechanism for the interaction between human CPSF30 and hFip1.
Hamilton, K., Tong, L.(2020) Genes Dev 34: 1753-1761
- PubMed: 33122294
- DOI: https://doi.org/10.1101/gad.343814.120
- Primary Citation of Related Structures:
7K95 - PubMed Abstract:
Most eukaryotic pre-mRNAs must undergo 3'-end cleavage and polyadenylation prior to their export from the nucleus. A large number of proteins in several complexes participate in this 3'-end processing, including cleavage and polyadenylation specificity factor (CPSF) in mammals. The CPSF30 subunit contains five CCCH zinc fingers (ZFs), with ZF2-ZF3 being required for the recognition of the AAUAAA poly(A) signal. ZF4-ZF5 recruits the hFip1 subunit of CPSF, although the details of this interaction have not been characterized. Here we report the crystal structure of human CPSF30 ZF4-ZF5 in complex with residues 161-200 of hFip1 at 1.9 Å resolution, illuminating the molecular basis for their interaction. Unexpectedly, the structure reveals one hFip1 molecule binding to each ZF4 and ZF5, with a conserved mode of interaction. Our mutagenesis studies confirm that the CPSF30-hFip1 complex has 1:2 stoichiometry in vitro. Mutation of each binding site in CPSF30 still allows one copy of hFip1 to bind, while mutation of both sites abrogates binding. Our fluorescence polarization binding assays show that ZF4 has higher affinity for hFip1, with a K d of 1.8 nM. We also demonstrate that two copies of the catalytic module of poly(A) polymerase (PAP) are recruited by the CPSF30-hFip1 complex in vitro, and both hFip1 binding sites in CPSF30 can support polyadenylation.
Organizational Affiliation:
Department of Biological Sciences, Columbia University, New York, New York 10027, USA.