Cell Surface Xyloglucan Recognition and Hydrolysis by the Human Gut Commensal Bacteroides uniformis.
Grondin, J.M., Dejean, G., Van Petegem, F., Brumer, H.(2022) Appl Environ Microbiol 88: e0156621-e0156621
- PubMed: 34731054
- DOI: https://doi.org/10.1128/AEM.01566-21
- Primary Citation of Related Structures:
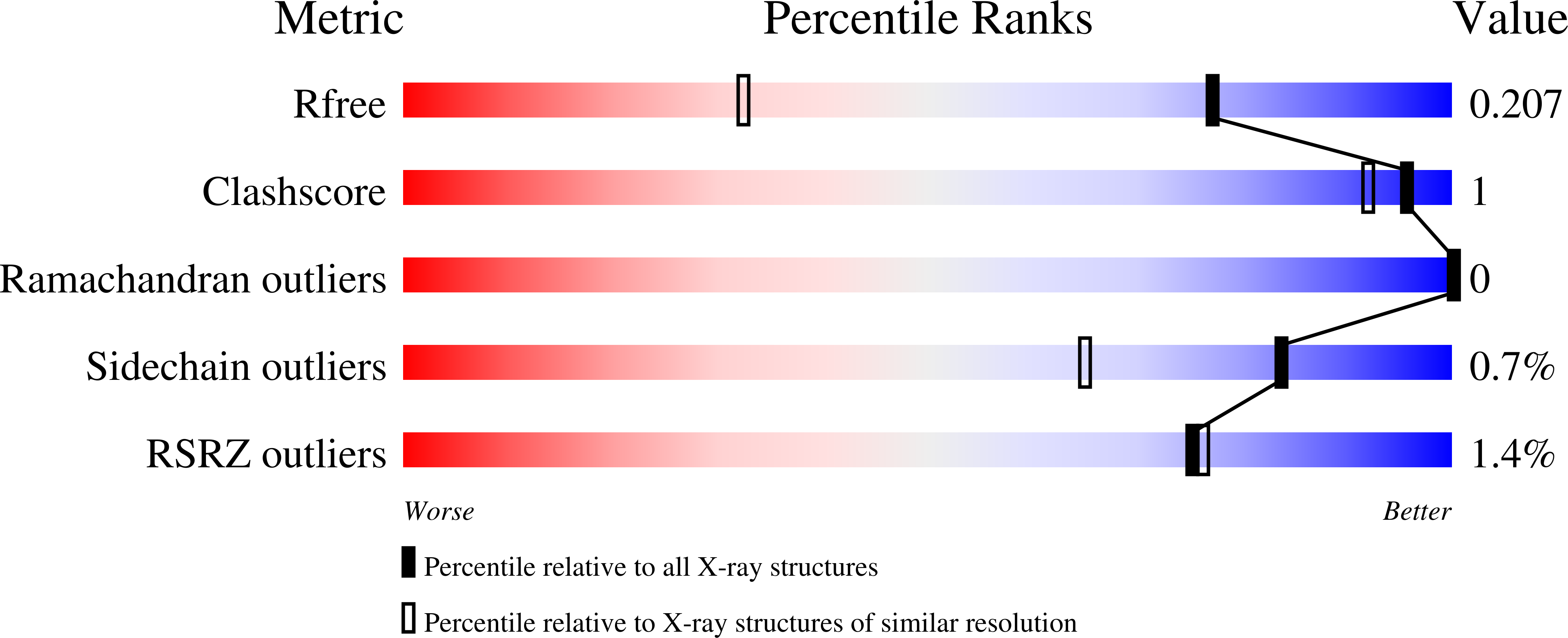
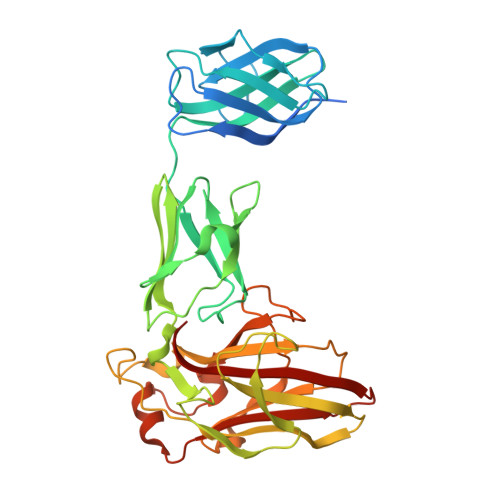
7K44 - PubMed Abstract:
Xyloglucan (XyG) is a ubiquitous plant cell wall hemicellulose that is targeted by a range of syntenic, microheterogeneous xyloglucan utilization loci (XyGUL) in Bacteroidetes species of the human gut microbiota (HGM), including Bacteroides ovatus and B. uniformis. Comprehensive biochemical and biophysical analyses have identified key differences in the protein complements of each locus that confer differential access to structurally diverse XyG side chain variants. A second, nonsyntenic XyGUL was previously identified in B. uniformis, although its function in XyG utilization compared to its syntenic counterpart was unclear. Here, complementary enzymatic product profiles and bacterial growth curves showcase the notable preference of Bu XyGUL2 surface glycan-binding proteins (SGBPs) to bind full-length XyG, as well as a range of oligosaccharides produced by the glycoside hydrolase family 5 (GH5_4) endo -xyloglucanase from this locus. We use isothermal titration calorimetry (ITC) to characterize this binding capacity and pinpoint the specific contributions of each protein to nutrient capture. The high-resolution structure of Bu XyGUL2 SGBP-B reveals remarkable putative binding site conservation with the canonical XyG-binding Bo XyGUL SGBP-B, supporting similar roles for these proteins in glycan capture. Together, these data underpin the central role of complementary XyGUL function in B. uniformis and broaden our systems-based and mechanistic understanding of XyG utilization in the HGM. IMPORTANCE The omnipresence of xyloglucans in the human diet has led to the evolution of heterogeneous gene clusters in several Bacteroidetes species in the HGM, each specially tuned to respond to the structural variations of these complex plant cell wall polysaccharides. Our research illuminates the complementary roles of syntenic and nonsyntenic XyGUL in B. uniformis in conferring growth on a variety of XyG-derived substrates, providing evidence of glycan-binding protein microadaptation within a single species. These data serve as a comprehensive overview of the binding capacities of the SGBPs from a nonsyntenic B. uniformis XyGUL and will inform future studies on the roles of complementary loci in glycan targeting by key HGM species.
Organizational Affiliation:
Michael Smith Laboratories, University of British Columbiagrid.17091.3e, Vancouver, British Columbia, Canada.