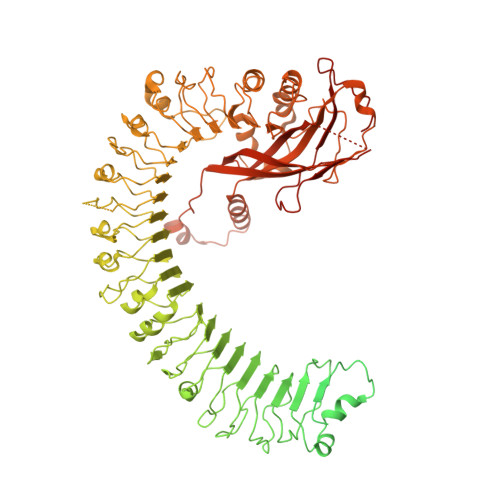
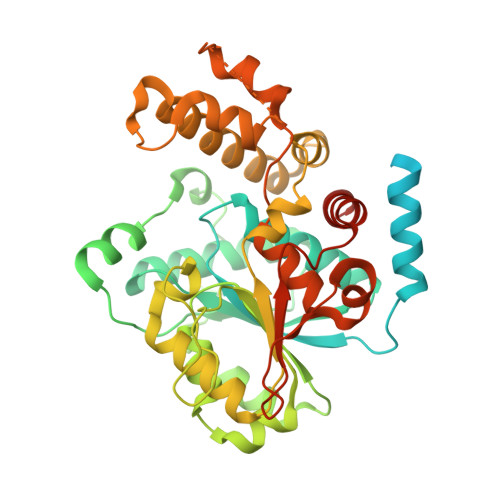
Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ.
Martin, R., Qi, T., Zhang, H., Liu, F., King, M., Toth, C., Nogales, E., Staskawicz, B.J.(2020) Science 370
- PubMed: 33273074
- DOI: https://doi.org/10.1126/science.abd9993
- Primary Citation of Related Structures:
7JLU, 7JLV, 7JLX - PubMed Abstract:
Plants and animals detect pathogen infection using intracellular nucleotide-binding leucine-rich repeat receptors (NLRs) that directly or indirectly recognize pathogen effectors and activate an immune response. How effector sensing triggers NLR activation remains poorly understood. Here we describe the 3.8-angstrom-resolution cryo-electron microscopy structure of the activated ROQ1 (recognition of XopQ 1), an NLR native to Nicotiana benthamiana with a Toll-like interleukin-1 receptor (TIR) domain bound to the Xanthomonas euvesicatoria effector XopQ ( Xanthomonas outer protein Q). ROQ1 directly binds to both the predicted active site and surface residues of XopQ while forming a tetrameric resistosome that brings together the TIR domains for downstream immune signaling. Our results suggest a mechanism for the direct recognition of effectors by NLRs leading to the oligomerization-dependent activation of a plant resistosome and signaling by the TIR domain.
Organizational Affiliation:
Biophysics Graduate Group, University of California, Berkeley, CA 94720, USA.