Crystal structures of PirA and PirB toxins from Photorhabdus akhurstii subsp. akhurstii K-1
Prashar, A., Kinkar, O.U., Kumar, A., Hadapad, A.B., Makde, R.D., Hire, R.S.(2023) Insect Biochem Mol Biol 162: 104014
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2023) Insect Biochem Mol Biol 162: 104014
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Insecticidal protein | 419 | Photorhabdus akhurstii | Mutation(s): 0 Gene Names: pirB | 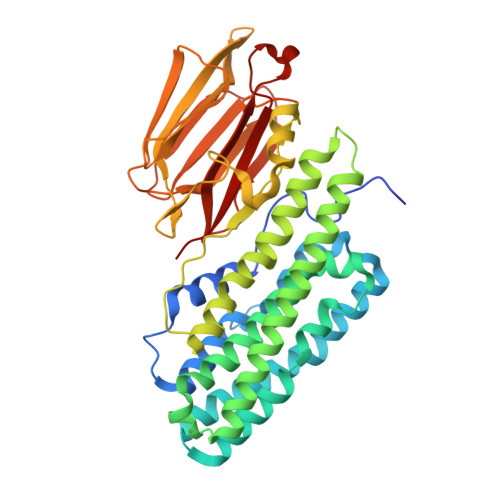 | |
UniProt | |||||
Find proteins for G5DBH4 (Photorhabdus akhurstii) Explore G5DBH4 Go to UniProtKB: G5DBH4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | G5DBH4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 73.802 | α = 90 |
| b = 89.274 | β = 90 |
| c = 124.657 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| PHENIX | model building |
| PHENIX | refinement |
| Coot | model building |