Revealing two important tryptophan residues with completely different roles in a dye-decolorizing peroxidase from Irpex lacteus F17.
Li, L., Wang, T., Chen, T., Huang, W., Zhang, Y., Jia, R., He, C.(2021) Biotechnol Biofuels 14: 128-128
- PubMed: 34059116
- DOI: https://doi.org/10.1186/s13068-021-01978-y
- Primary Citation of Related Structures:
7D8M - PubMed Abstract:
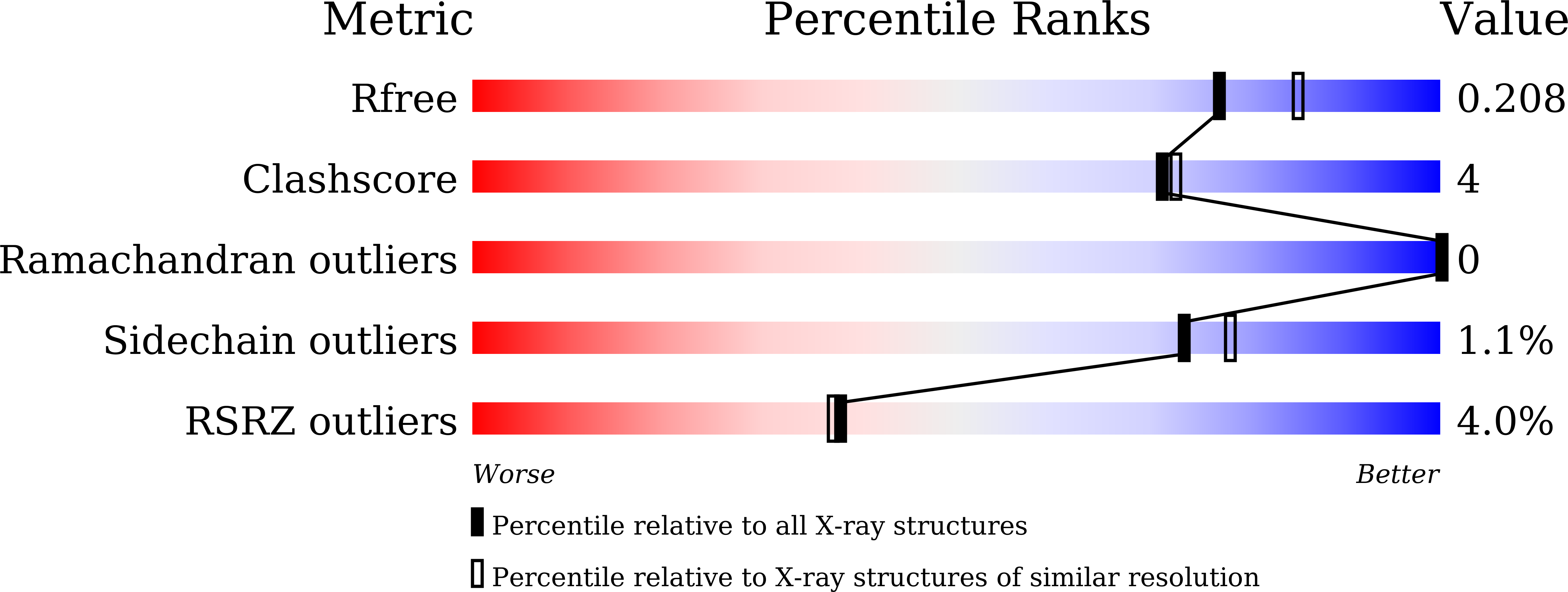
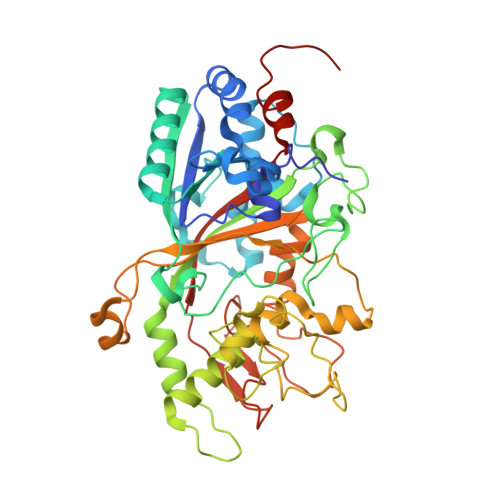
Dye-decolorizing peroxidases (DyPs) represent a novel family of heme peroxidases that use H 2 O 2 as the final electron acceptor to catalyze the oxidation of various organic compounds. A DyP from Irpex lacteus F17 (Il-DyP4, corresponding to GenBank MG209114), obtained by heterologous expression, exhibits a high catalytic efficiency for phenolic compounds and a strong decolorizing ability toward various synthetic dyes. However, the enzyme structure and the catalytic residues involved in substrate oxidation remain poorly understood. Here, we obtained a high-resolution structure (2.0 Å, PDB: 7D8M) of Il‑DyP4 with α-helices, anti-parallel β-sheets and one ferric heme cofactor sandwiched between two domains. The crystal structure of Il‑DyP4 revealed two heme access channels leading from the enzyme molecular surface to its heme region, and also showed four conserved amino acid residues forming the pocket for the conversion of hydrogen peroxide into the water molecule. In addition, we found that Trp264 and Trp380, were two important residues with different roles in Il‑DyP4, by using site-directed mutagenesis and an electron paramagnetic resonance (EPR) study. Trp264 is a noncatalytic residue that mainly is used for maintaining the normal spatial conformation of the heme region and the high-spin state of heme Fe 3+ of Il‑DyP4, while Trp380 serves as the surface-exposed radical-forming residue that is closely related to the oxidation of substrates including not only bulky dyes, but also simple phenols. This study is important for better understanding the catalytic properties of fungal DyPs and their structure-function relationships.
Organizational Affiliation:
School of Life Science, Economic and Technology Development Zone, Anhui University, 111 jiulong Road, Hefei, Anhui, PR China, 230601.