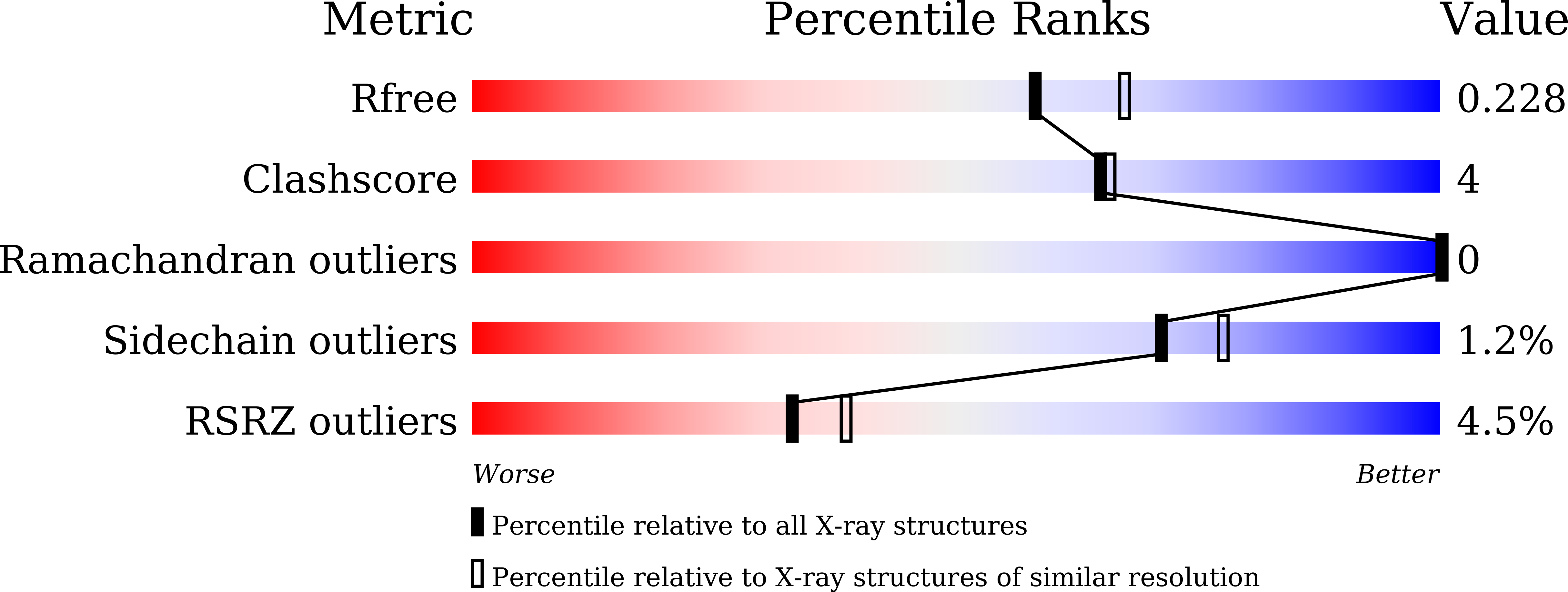
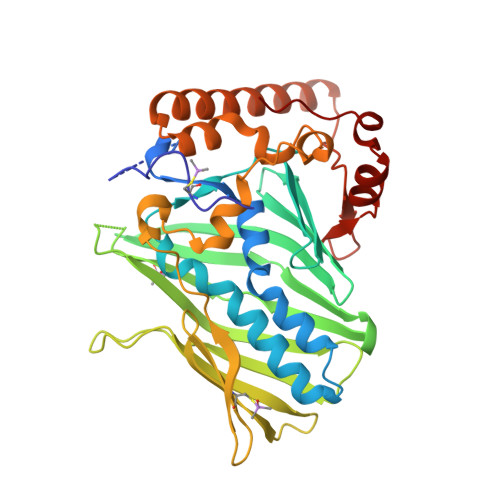
The crystal structure of ORP3 reveals the conservative PI4P binding pattern.
Dong, X., Wang, Z., Ye, S., Zhang, R.(2020) Biochem Biophys Res Commun 529: 1005-1010
- PubMed: 32819557
- DOI: https://doi.org/10.1016/j.bbrc.2020.06.090
- Primary Citation of Related Structures:
7CYZ - PubMed Abstract:
Oxysterol-binding protein (OSBP) and its related protein (ORP) constitute a conserved family of lipid transfer proteins (LTPs). ORPs have been implicated as intracellular lipid exchanger and sensor in recent years, which regulate the lipid homeostasis and signal pathway. OSBP-related protein 3 plays key role in controlling cell adhesion and migration and could be developed as the drug target for cancer therapy. Here, we report the crystal structures of human ORP3 ORD to 2.1 Å and ORD-PI4P complex to 3.2 Å. The binding assay in vitro confirms the ORP3 has the capability of PI4P binding. This study further verifies that the PI4P is the common ligand of all ORPs and ORPs should be the lipid exchanger in membrane contact sites(MCS).
Organizational Affiliation:
National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, 100101, People's Republic of China. Electronic address: dongxuedejia@hotmail.com.