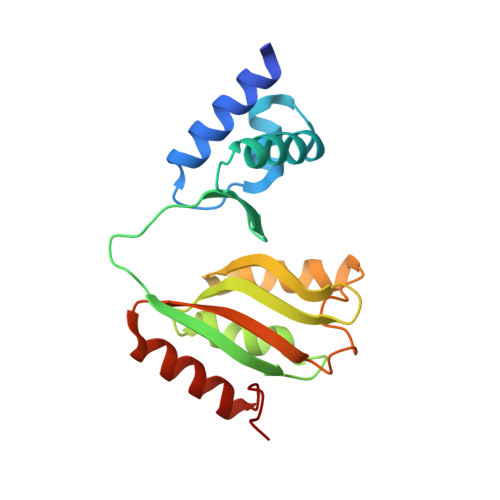
Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Lee, D.W., Park, Y.W., Lee, M.Y., Jeong, K.H., Lee, J.Y.(2020) Sci Rep 10: 21039-21039
- PubMed: 33273654
- DOI: https://doi.org/10.1038/s41598-020-78148-x
- Primary Citation of Related Structures:
7CV0, 7CV2 - PubMed Abstract:
The niacin-responsive repressor, NiaR, is transcriptional repressor of certain nicotinamide adenine dinucleotide (NAD) biosynthetic genes in response to an increase in niacin levels. NAD is a vital molecule involved in various cellular redox reactions as an electron donor or electron acceptor. The NiaR family is conserved broadly in the Bacillus/Clostridium group, as well as in the Fusobacteria and Thermotogales lineages. The NiaR structure consists of two domains: an N-terminal DNA-binding domain, and a C-terminal regulation domain containing a metal-binding site. In this paper, we report the crystal structures of apo and niacin-bound forms of NiaR from Bacillus halodurans (BhNiaR). The analysis of metal-binding and niacin-binding sites through the apo and niacin-bound structures is described. Each N- and C-terminal domain structure of BhNiaR is almost identical with NiaR from Thermotoga maritima, but the overall domain arrangement is quite different. A zinc ion is fully occupied in each subunit with well-conserved residues in the C-terminal domain. Niacin is also located at a hydrophobic pocket near the zinc ion in the C-terminal domain.
Organizational Affiliation:
Department of Life Science, Dongguk University-Seoul, Ilsandong-gu, Goyang-si, Gyeonggi-do, 10326, Republic of Korea.