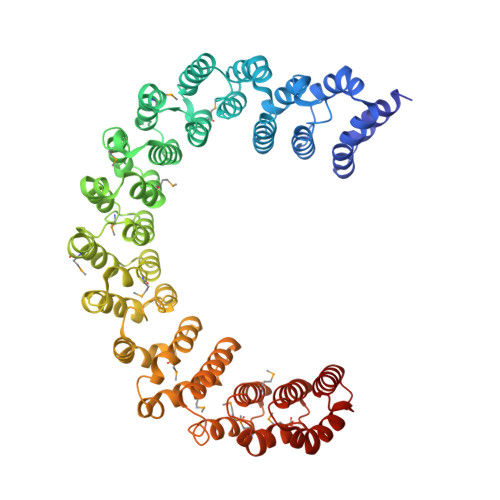Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase.
Zheng, H., Qi, Y., Hu, S., Cao, X., Xu, C., Yin, Z., Chen, X., Li, Y., Liu, W., Li, J., Wang, J., Wei, G., Liang, K., Chen, F.X., Xu, Y.(2020) Science 370
- PubMed: 33243860
- DOI: https://doi.org/10.1126/science.abb5872
- Primary Citation of Related Structures:
7CUN - PubMed Abstract:
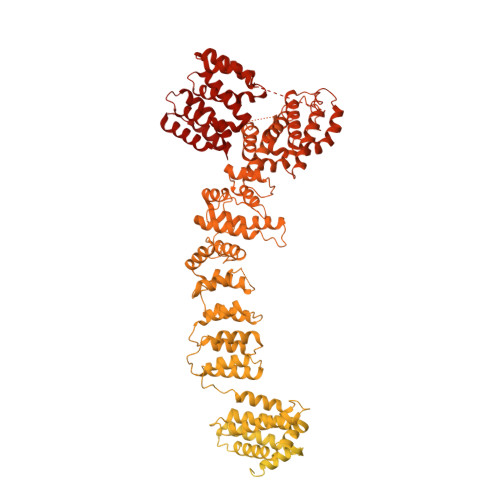
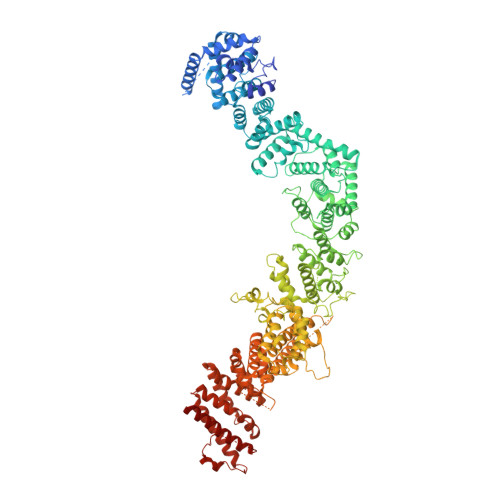
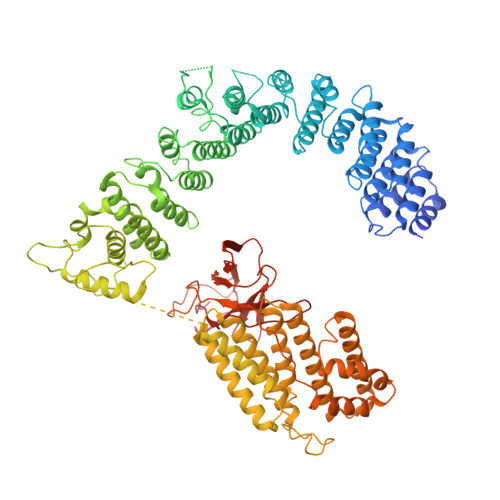
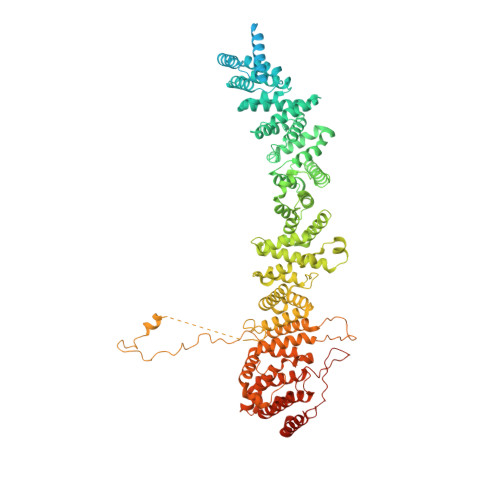
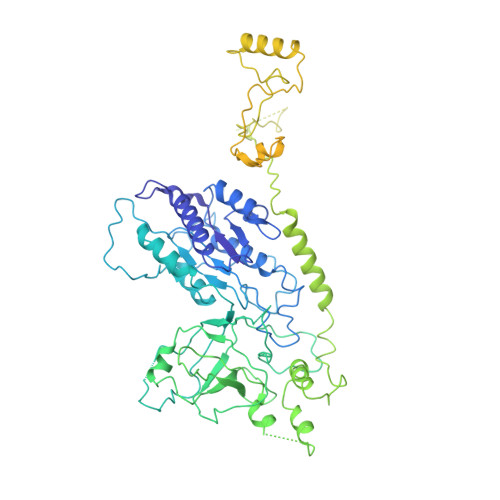
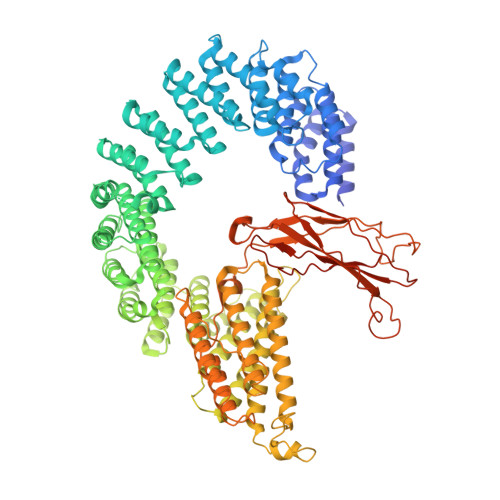
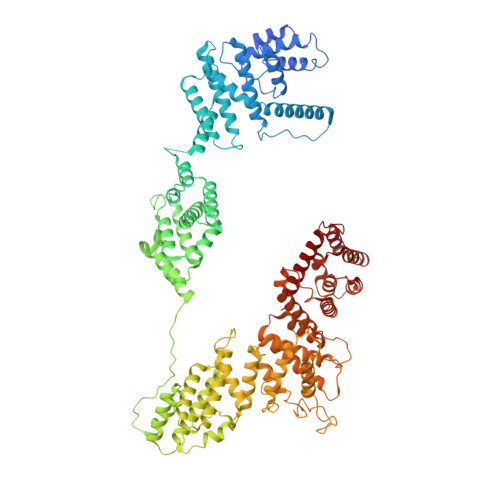
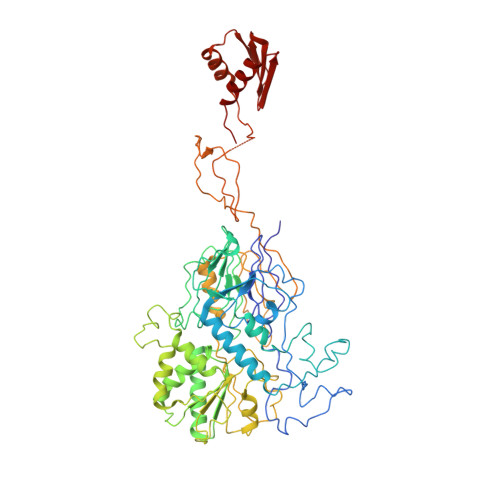
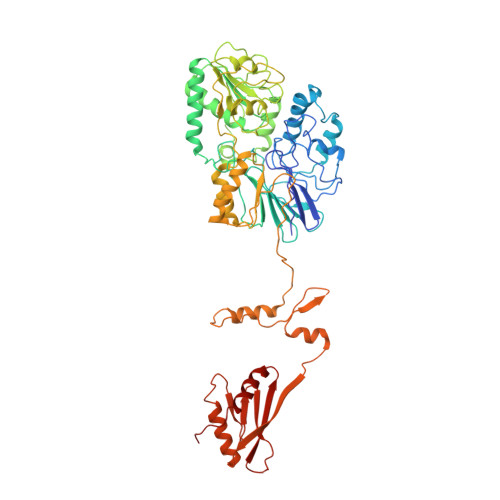
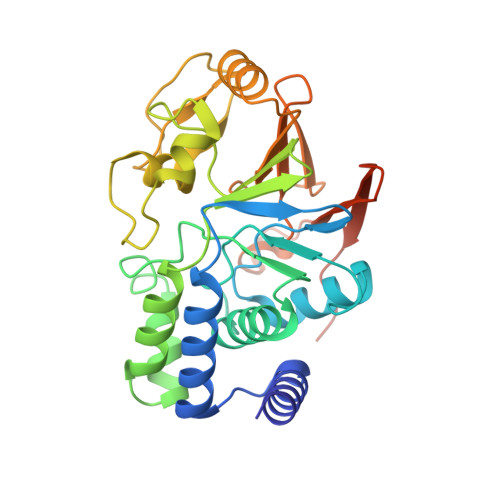
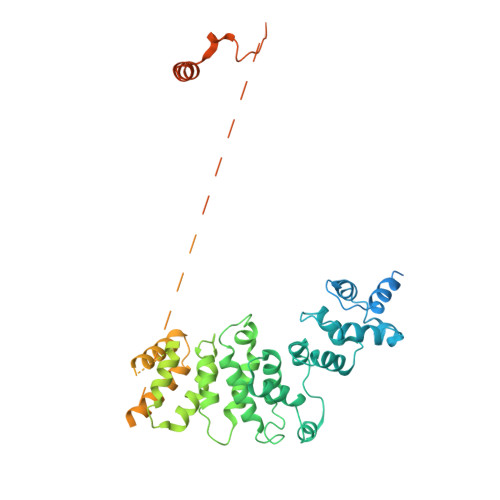
The 14-subunit metazoan-specific Integrator contains an endonuclease that cleaves nascent RNA transcripts. Here, we identified a complex containing Integrator and protein phosphatase 2A core enzyme (PP2A-AC), termed INTAC. The 3.5-angstrom-resolution structure reveals that nine human Integrator subunits and PP2A-AC assemble into a cruciform-shaped central scaffold formed by the backbone and shoulder modules, with the phosphatase and endonuclease modules flanking the opposite sides. As a noncanonical PP2A holoenzyme, the INTAC complex dephosphorylates the carboxy-terminal repeat domain of RNA polymerase II at serine-2, -5, and -7 and thus regulates transcription. Our study extends the function of PP2A to transcriptional regulation and reveals how dual enzymatic activities-RNA cleavage and RNA polymerase II dephosphorylation-are structurally and functionally integrated into the INTAC complex.
Organizational Affiliation:
Fudan University Shanghai Cancer Center, Institutes of Biomedical Sciences, State Key Laboratory of Genetic Engineering and Shanghai Key Laboratory of Medical Epigenetics, Shanghai Medical College of Fudan University, Shanghai 200032, China.