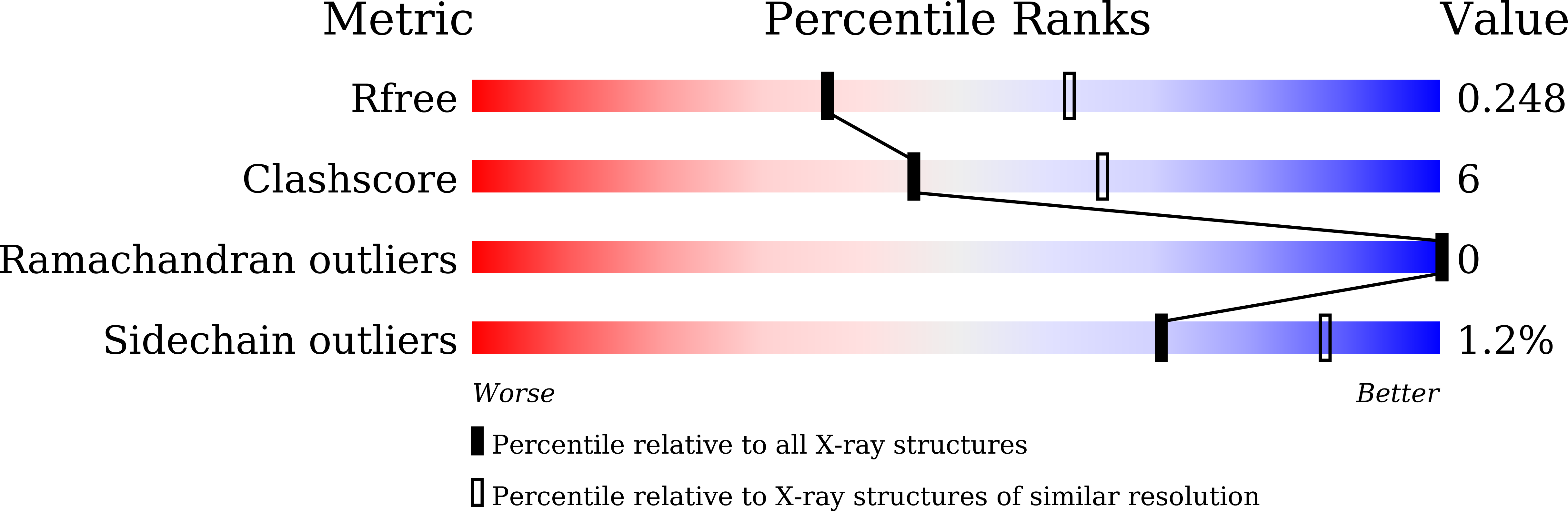
The Crystal Structure of the MHC Class I (MHC-I) Molecule in the Green Anole Lizard Demonstrates the Unique MHC-I System in Reptiles.
Wang, Y., Qu, Z., Ma, L., Wei, X., Zhang, N., Zhang, B., Xia, C.(2021) J Immunol 206: 1653-1667
- PubMed: 33637616
- DOI: https://doi.org/10.4049/jimmunol.2000992
- Primary Citation of Related Structures:
7CPO - PubMed Abstract:
The reptile MHC class I (MCH-I) and MHC class II proteins are the key molecules in the immune system; however, their structure has not been investigated. The crystal structure of green anole lizard peptide-MHC-I-β2m (pMHC-I or p Anca -UA*0101) was determined in the current study. Subsequently, the features of p Anca -UA*0101 were analyzed and compared with the characteristics of pMHC-I of four classes of vertebrates. The amino acid sequence identities between Anca -UA*0101 and MHC-I from other species are <50%; however, the differences between the species were reflected in the topological structure. Significant characteristics of p Anca -UA*0101 include a specific flip of ∼88° and an upward shift adjacent to the C terminus of the α1- and α2-helical regions, respectively. Additionally, the lizard MHC-I molecule has an insertion of 2 aa (VE) at positions 55 and 56. The pushing force from 55-56VE triggers the flip of the α1 helix. Mutagenesis experiments confirmed that the 55-56VE insertion in the α1 helix enhances the stability of p Anca -UA*0101. The peptide presentation profile and motif of p Anca -UA*0101 were confirmed. Based on these results, the proteins of three reptile lizard viruses were used for the screening and confirmation of the candidate epitopes. These data enhance our understanding of the systematic differences between five classes of vertebrates at the gene and protein levels, the formation of the pMHC-I complex, and the evolution of the MHC-I system.
Organizational Affiliation:
Department of Microbiology and Immunology, College of Veterinary Medicine, China Agricultural University, Beijing 100193, China.