Structural mechanism of bivalent histone H3K4me3K9me3 recognition by the Spindlin1/C11orf84 complex in rRNA transcription activation.
Du, Y., Yan, Y., Xie, S., Huang, H., Wang, X., Ng, R.K., Zhou, M.M., Qian, C.(2021) Nat Commun 12: 949-949
- PubMed: 33574238
- DOI: https://doi.org/10.1038/s41467-021-21236-x
- Primary Citation of Related Structures:
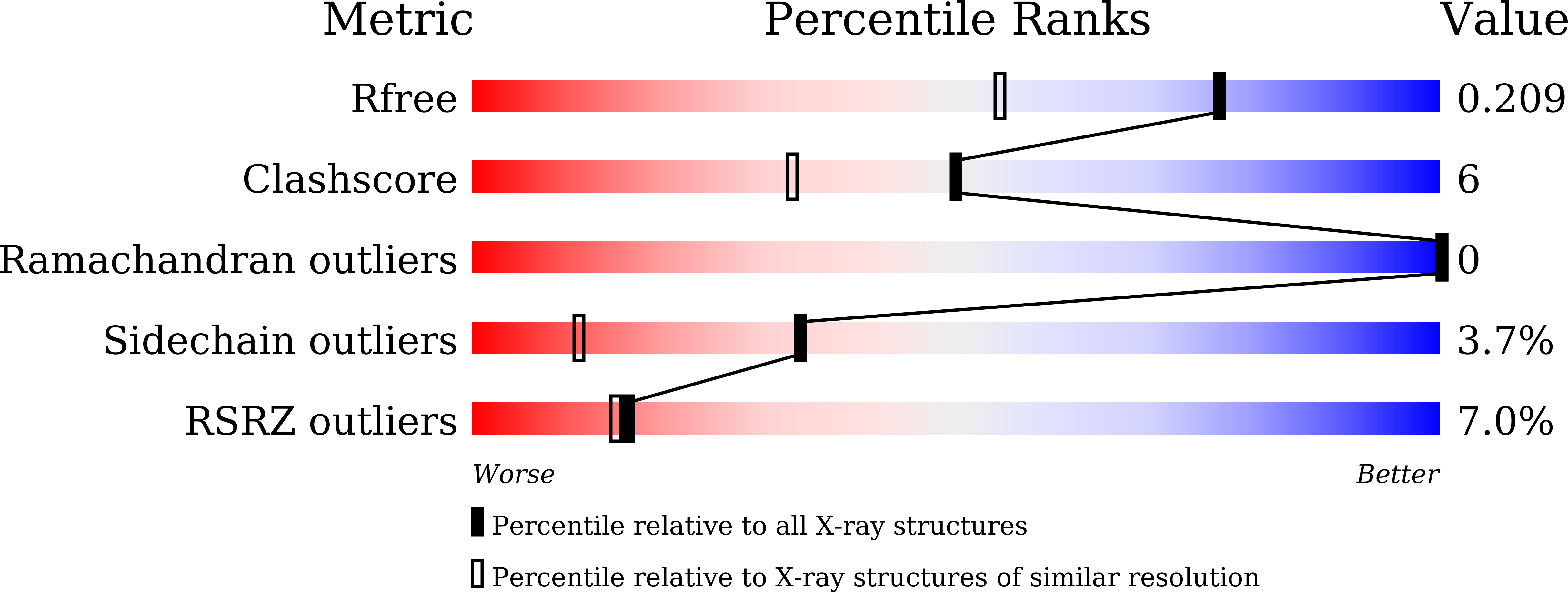
7CNA - PubMed Abstract:
Spindlin1 is a unique multivalent epigenetic reader that facilitates ribosomal RNA transcription. In this study, we provide molecular and structural basis by which Spindlin1 acts in complex with C11orf84 to preferentially recognize non-canonical bivalent mark of trimethylated lysine 4 and lysine 9 present on the same histone H3 tail (H3K4me3K9me3). We demonstrate that C11orf84 binding stabilizes Spindlin1 and enhances its association with bivalent H3K4me3K9me3 mark. The functional analysis suggests that Spindlin1/C11orf84 complex can displace HP1 proteins from H3K4me3K9me3-enriched rDNA loci, thereby facilitating the conversion of these poised rDNA repeats from the repressed state to the active conformation, and the consequent recruitment of RNA Polymerase I for rRNA transcription. Our study uncovers a previously unappreciated mechanism of bivalent H3K4me3K9me3 recognition by Spindlin1/C11orf84 complex required for activation of rRNA transcription.
Organizational Affiliation:
School of Biomedical Sciences, The University of Hong Kong, Hong Kong Island, Hong Kong.