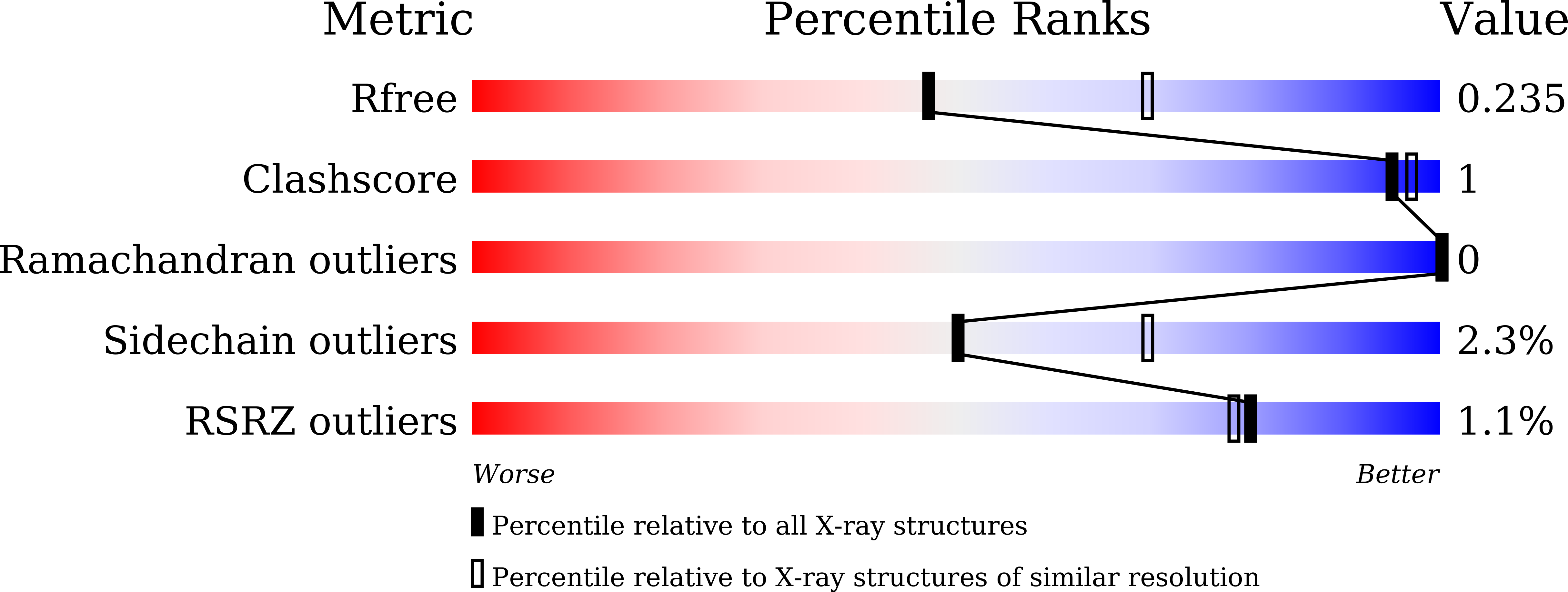
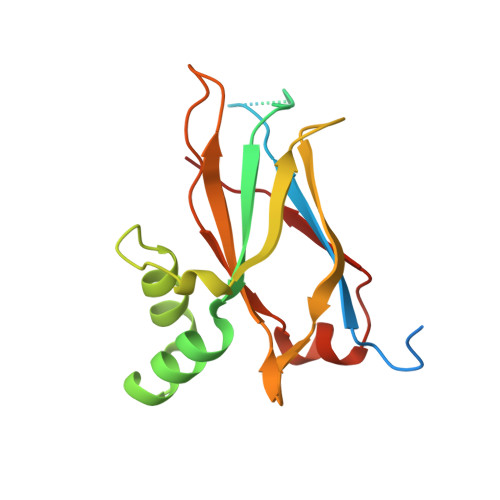
Crystal structure of the atypically adhesive SpaB basal pilus subunit: Mechanistic insights about its incorporation in lactobacillar SpaCBA pili.
Megta, A.K., Pratap, S., Kant, A., Palva, A., von Ossowski, I., Krishnan, V.(2020) Curr Res Struct Biol 2: 229-238