Structural and Biochemical Analysis Reveals a Distinct Catalytic Site of Salicylate 5-Monooxygenase NagGH from Rieske Dioxygenases.
Hou, Y.J., Guo, Y., Li, D.F., Zhou, N.Y.(2021) Appl Environ Microbiol 87
- PubMed: 33452034
- DOI: https://doi.org/10.1128/AEM.01629-20
- Primary Citation of Related Structures:
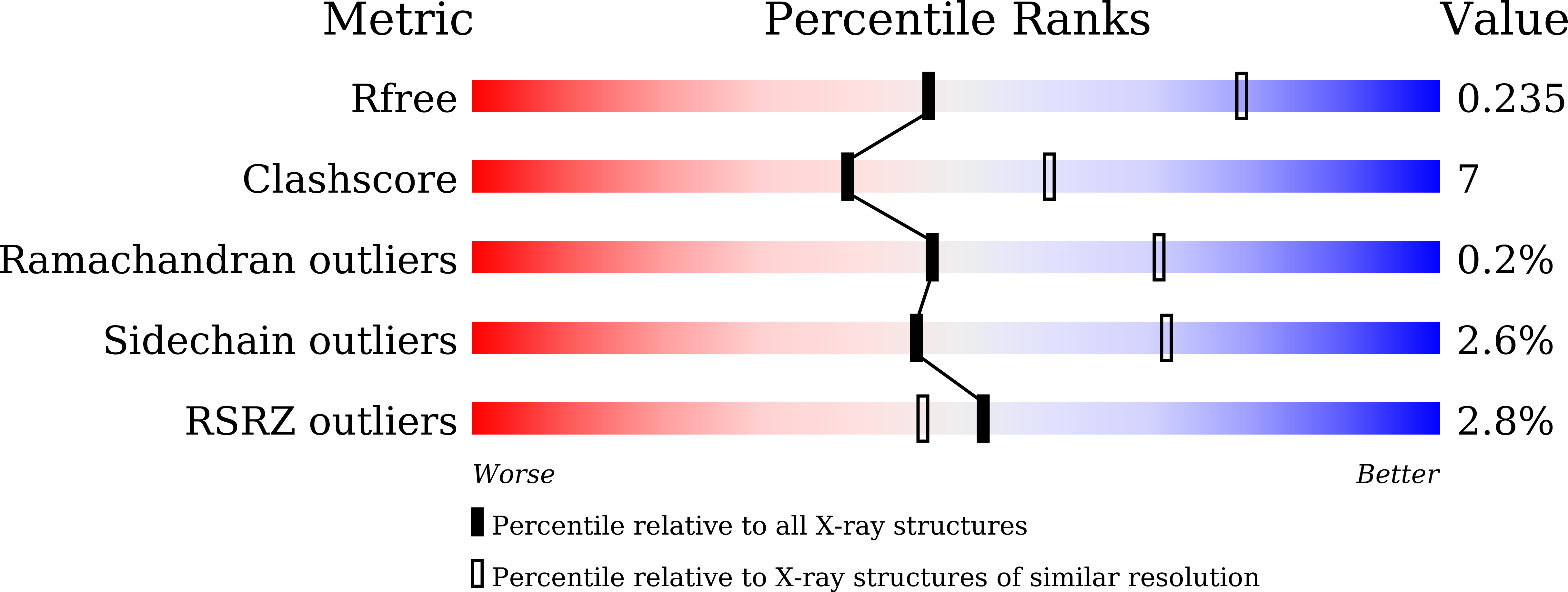
7C8Z - PubMed Abstract:
Rieske nonheme iron oxygenases (ROs) catalyze the oxidation of a wide variety of substrates and play important roles in aromatic compound degradation and polycyclic aromatic hydrocarbon degradation. Those Rieske dioxygenases that usually act on hydrophobic substrates have been extensively studied and structurally characterized. Here, we report the crystal structure of a novel Rieske monooxygenase, NagGH, the oxygenase component of a salicylate 5-monooxygenase from Ralstonia sp. strain U2 that catalyzes the hydroxylation of a hydrophilic substrate salicylate (2-hydroxybenzoate), forming gentisate (2, 5-dihydroxybenzoate). The large subunit NagG and small subunit NagH share the same fold as that for their counterparts of Rieske dioxygenases and assemble the same α 3 β 3 hexamer, despite that they share low (or no identity for NagH) sequence identities with these dioxygenase counterparts. A potential substrate-binding pocket was observed in the vicinity of the nonheme iron site. It featured a positively charged residue Arg323 that was surrounded by hydrophobic residues. The shift of nonheme iron atom caused by residue Leu228 disrupted the usual substrate pocket observed in other ROs. Residue Asn218 at the usual substrate pocket observed in other ROs was likewise involved in substrate binding and oxidation, yet residues Gln316 and Ser367, away from the usual substrate pocket of other ROs, were shown to play a more important role in substrate oxidation than Asn218. The unique binding pocket and unusual substrate-protein hydrophilic interaction provide new insights into Rieske monooxygenases. IMPORTANCE Rieske oxygenases are involved in the degradation of various aromatic compounds. These dioxygenases usually carry out hydroxylation of hydrophobic aromatic compounds and supply substrates with hydroxyl groups for extradiol/intradiol dioxygenases to cleave rings, and have been extensively studied. Salicylate 5-hydroxylase NagGH is a novel Rieske monooxygenase with high similarity to Rieske dioxygenases, and also shares reductase and ferredoxin similarity with a Rieske dioxygenase naphthalene 1,2-dioxygenase (NagAcAd) in Ralstonia sp. strain U2. The structure of NagGH, the oxygenase component of salicylate 5-monooxygenase, gives a representative of those monooxygenases and will help us understand the mechanism of their substrate binding and product regio-selectivity.
Organizational Affiliation:
State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.