Modulation of Lymphocyte Potassium Channel KV1.3 by Membrane-Penetrating, Joint-Targeting Immunomodulatory Plant Defensin.
Ong, S.T., Bajaj, S., Tanner, M.R., Chang, S.C., Krishnarjuna, B., Ng, X.R., Morales, R.A.V., Chen, M.W., Luo, D., Patel, D., Yasmin, S., Ng, J.J.H., Zhuang, Z., Nguyen, H.M., El Sahili, A., Lescar, J., Patil, R., Charman, S.A., Robins, E.G., Goggi, J.L., Tan, P.W., Sadasivam, P., Ramasamy, B., Hartimath, S.V., Dhawan, V., Bednenko, J., Colussi, P., Wulff, H., Pennington, M.W., Kuyucak, S., Norton, R.S., Beeton, C., Chandy, K.G.(2020) Acs Pharmacol Transl Sci 3: 720-736
- PubMed: 32832873
- DOI: https://doi.org/10.1021/acsptsci.0c00035
- Primary Citation of Related Structures:
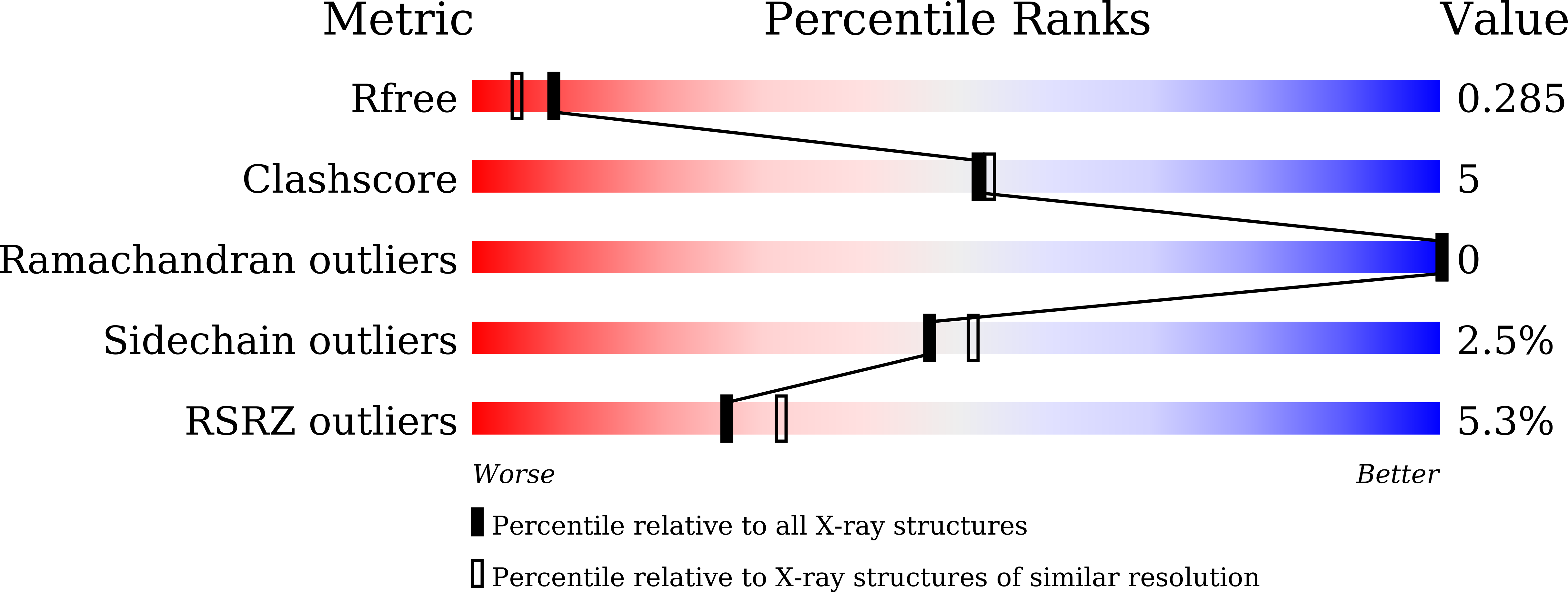
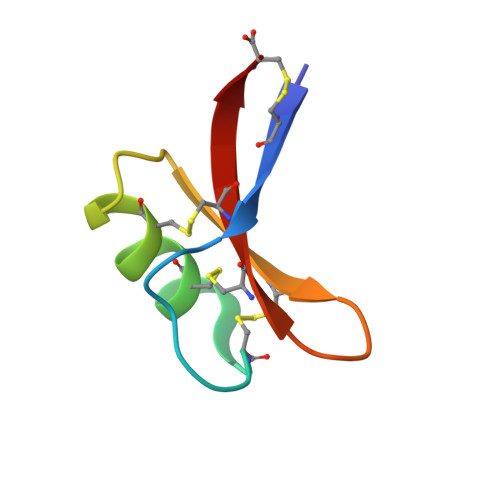
7C2P, 7C31 - PubMed Abstract:
We describe a cysteine-rich, membrane-penetrating, joint-targeting, and remarkably stable peptide, EgK5, that modulates voltage-gated K V 1.3 potassium channels in T lymphocytes by a distinctive mechanism. EgK5 enters plasma membranes and binds to K V 1.3, causing current run-down by a phosphatidylinositol 4,5-bisphosphate-dependent mechanism. EgK5 exhibits selectivity for K V 1.3 over other channels, receptors, transporters, and enzymes. EgK5 suppresses antigen-triggered proliferation of effector memory T cells, a subset enriched among pathogenic autoreactive T cells in autoimmune disease. PET-CT imaging with 18 F-labeled EgK5 shows accumulation of the peptide in large and small joints of rodents. In keeping with its arthrotropism, EgK5 treats disease in a rat model of rheumatoid arthritis. It was also effective in treating disease in a rat model of atopic dermatitis. No signs of toxicity are observed at 10-100 times the in vivo dose. EgK5 shows promise for clinical development as a therapeutic for autoimmune diseases.
Organizational Affiliation:
Lee Kong Chian School of Medicine, Nanyang Technological University Singapore, Experimental Medicine Building, 59 Nanyang Drive, Singapore 636921.