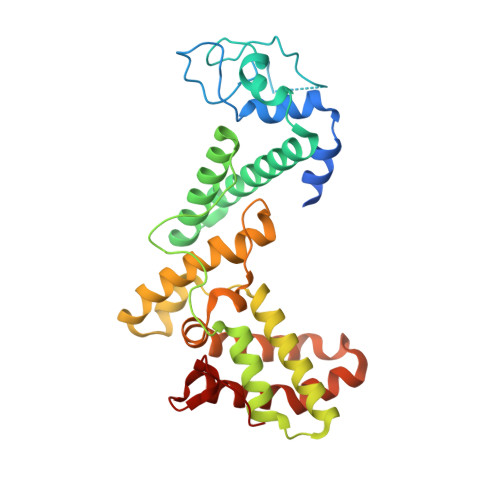
Crystal structure of GCN5 PCAF N-terminal domain reveals atypical ubiquitin ligase structure.
Toma-Fukai, S., Hibi, R., Naganuma, T., Sakai, M., Saijo, S., Shimizu, N., Matsumoto, M., Shimizu, T.(2020) J Biol Chem 295: 14630-14639
- PubMed: 32820047
- DOI: https://doi.org/10.1074/jbc.RA120.013431
- Primary Citation of Related Structures:
7BY1 - PubMed Abstract:
General control nonderepressible 5 (GCN5, also known as Kat2a) and p300/CBP-associated factor (PCAF, also known as Kat2b) are two homologous acetyltransferases. Both proteins share similar domain architecture consisting of a PCAF N-terminal (PCAF_N) domain, acetyltransferase domain, and a bromodomain. PCAF also acts as a ubiquitin E3 ligase whose activity is attributable to the PCAF_N domain, but its structural aspects are largely unknown. Here, we demonstrated that GCN5 exhibited ubiquitination activity in a similar manner to PCAF and its activity was supported by the ubiquitin-conjugating enzyme UbcH5. Moreover, we determined the crystal structure of the PCAF_N domain at 1.8 Å resolution and found that PCAF_N domain folds into a helical structure with a characteristic binuclear zinc region, which was not predicted from sequence analyses. The zinc region is distinct from known E3 ligase structures, suggesting this region may form a new class of E3 ligase. Our biochemical and structural study provides new insight into not only the functional significance of GCN5 but also into ubiquitin biology.
Organizational Affiliation:
Graduate School of Pharmaceutical Sciences, The University of Tokyo, Bunkyo-ku, Tokyo, Japan.