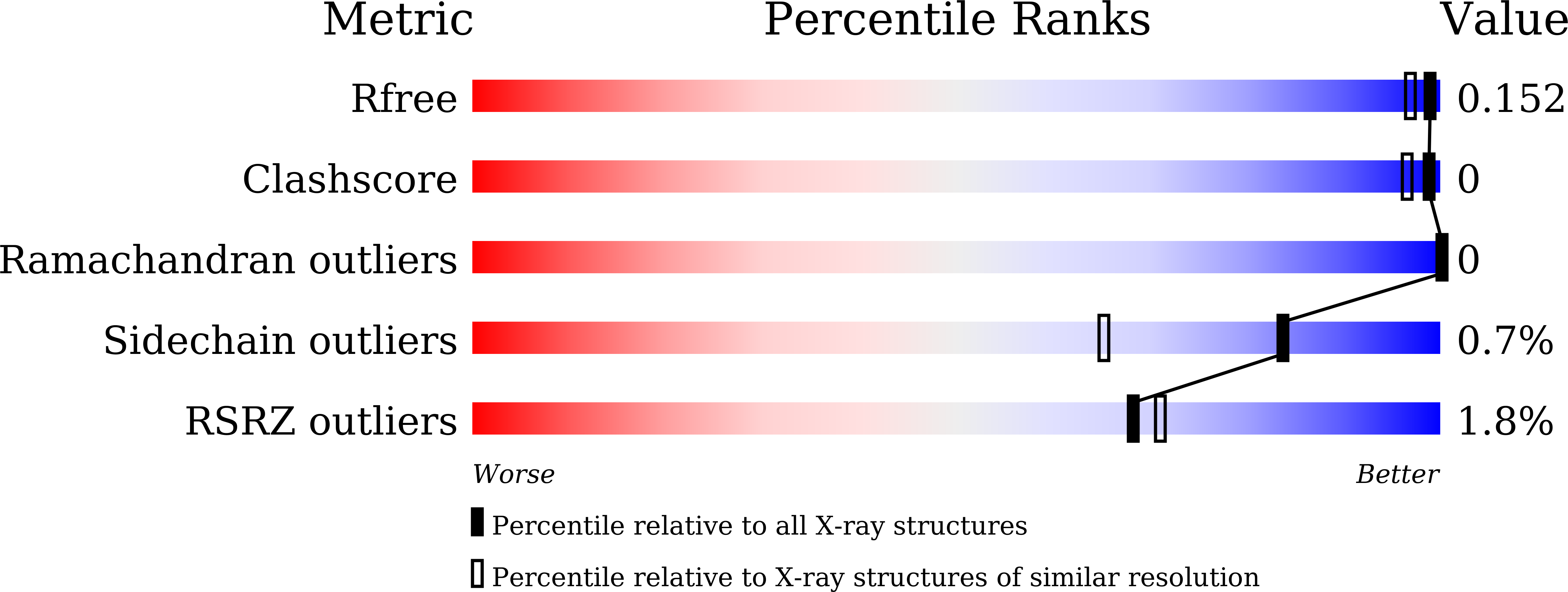
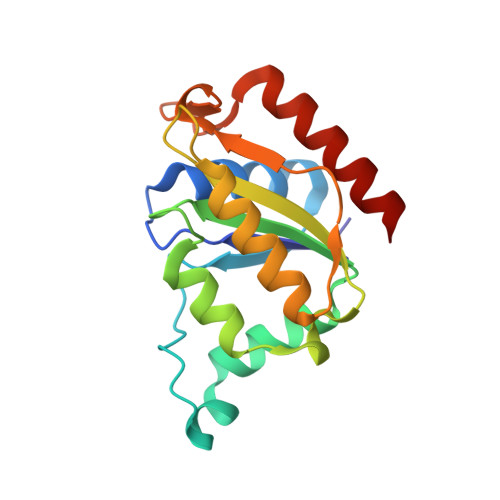
Purification, characterization, and crystal structure of YhdA-type azoreductase from Bacillus velezensis.
Bafana, A., Khan, F., Suguna, K.(2021) Proteins 89: 483-492
- PubMed: 33289153
- DOI: https://doi.org/10.1002/prot.26032
- Primary Citation of Related Structures:
7BX9 - PubMed Abstract:
Azoreductases are being extensively investigated for their ability to initiate degradation of recalcitrant azo dyes through reduction of azo bonds. There is great interest in studying their diversity, structure, and function to facilitate better understanding and effective application. Current study reports azoreductase enzyme from Bacillus velezensis, which showed 69.5% identity to the Bacillus subtilis azoreductase YhdA. The enzyme was homotetrameric and molecular weight of each subunit was 20 kDa. It decolorized azo dyes with different structures. The V max for decolorization of congo red, methyl orange and methyl red was 14.7, 28.6, and 77.9 nmol/min/mg, respectively. The enzyme contained FMN as cofactor and used NADPH as the favored co-substrate. It was oxygen-insensitive, but the presence of reducing agents enhanced its activity, which is a new finding. The azoreductase expression in B. velezensis was found to be unaffected by addition of azo dyes, although azo dyes are known to induce azoreductase expression in few organisms. The enzyme was thermostable with melting temperature of 89.5°C and functioned in wide temperature range. Further, the enzyme was crystallized and its structure was solved. The structural basis of its functional attributes is discussed. In our knowledge, this is the first report on characterization of azoreductase enzyme from B. velezensis.
Organizational Affiliation:
Director's Research Cell, CSIR-NEERI (National Environmental Engineering Research Institute), Nagpur, India.