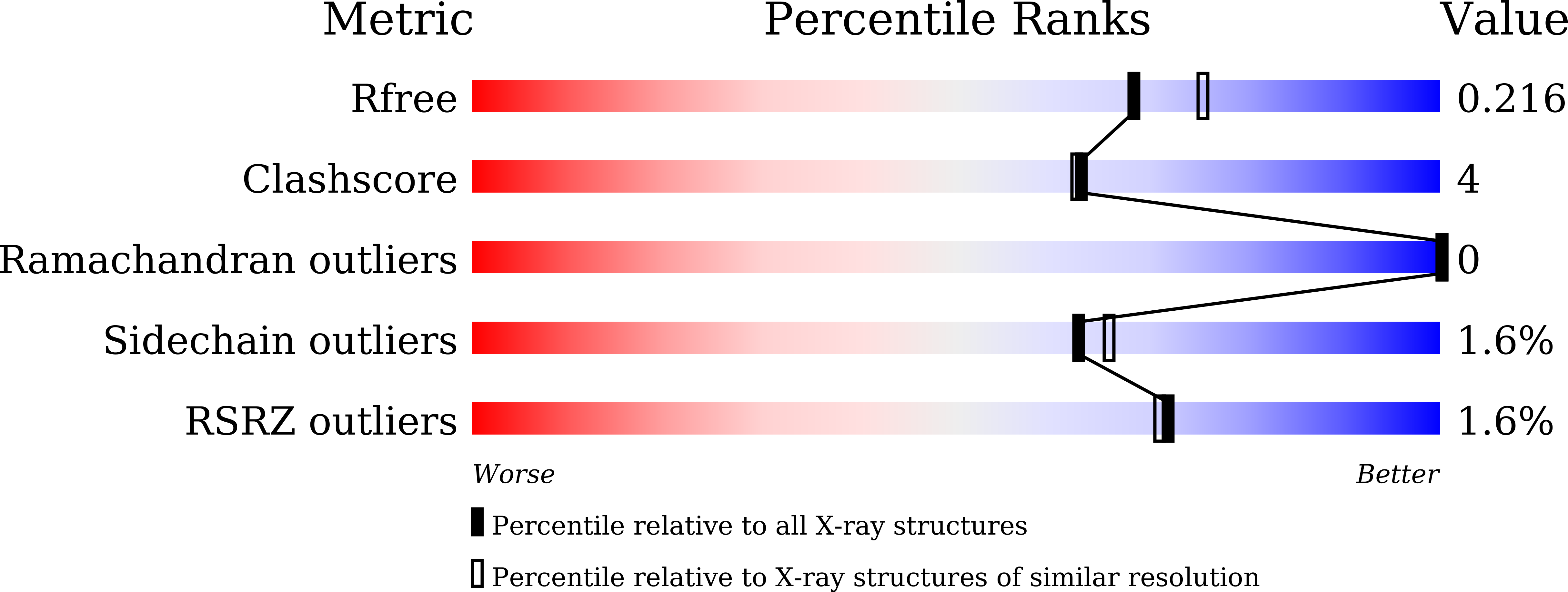
An Ice-Binding Protein from an Antarctic Ascomycete Is Fine-Tuned to Bind to Specific Water Molecules Located in the Ice Prism Planes.
Yamauchi, A., Arai, T., Kondo, H., Sasaki, Y.C., Tsuda, S.(2020) Biomolecules 10
- PubMed: 32414092
- DOI: https://doi.org/10.3390/biom10050759
- Primary Citation of Related Structures:
7BWX, 7BWY - PubMed Abstract:
Many microbes that survive in cold environments are known to secrete ice-binding proteins (IBPs). The structure-function relationship of these proteins remains unclear. A microbial IBP denoted Anp IBP was recently isolated from a cold-adapted fungus, Antarctomyces psychrotrophicus . The present study identified an orbital illumination (prism ring) on a globular single ice crystal when soaked in a solution of fluorescent Anp IBP, suggesting that Anp IBP binds to specific water molecules located in the ice prism planes. In order to examine this unique ice-binding mechanism, we carried out X-ray structural analysis and mutational experiments. It appeared that Anp IBP is made of 6-ladder β-helices with a triangular cross section that accompanies an "ice-like" water network on the ice-binding site. The network, however, does not exist in a defective mutant. Anp IBP has a row of four unique hollows on the IBS, where the distance between the hollows (14.7 Å) is complementary to the oxygen atom spacing of the prism ring. These results suggest the structure of Anp IBP is fine-tuned to merge with the ice-water interface of an ice crystal through its polygonal water network and is then bound to a specific set of water molecules constructing the prism ring to effectively halt the growth of ice.
Organizational Affiliation:
Graduate School of Life Science, Hokkaido University, Sapporo 060-0810, Japan.