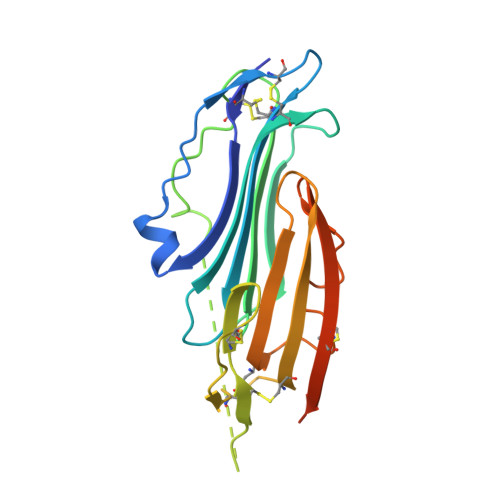
Crystal structure of TEX101, a glycoprotein essential for male fertility, reveals the presence of tandemly arranged Ly6/uPAR domains.
Masutani, M., Sakurai, S., Shimizu, T., Ohto, U.(2020) FEBS Lett 594: 3020-3031
- PubMed: 32608065
- DOI: https://doi.org/10.1002/1873-3468.13875
- Primary Citation of Related Structures:
7BPR, 7BPS - PubMed Abstract:
Testis-expressed gene 101 (TEX101) is a glycosyl-phosphatidylinositol-anchored glycoprotein essential for sperm fertility and spermatogenesis. TEX101 interacts with lymphocyte antigen 6 complex, locus K (Ly6k) as well as a disintegrin and metallopeptidase domain 3 (ADAM3). Although these proteins are considered essential for fertility, the associated mechanisms remain uncharacterized. Herein, we determined the crystal structure of human and mouse TEX101, revealing that TEX101 contains two tandem Ly6/uPAR (LU) domains. Detailed structural analyses revealed characteristic surfaces of TEX101 that may be involved in the interactions with other proteins or membranes. These results provide the structural basis for the role of TEX101 in fertilization and could contribute to developing diagnostic methods and treatments for infertility or developing male contraceptives.
Organizational Affiliation:
Graduate School of Pharmaceutical Sciences, The University of Tokyo, Hongo, Japan.