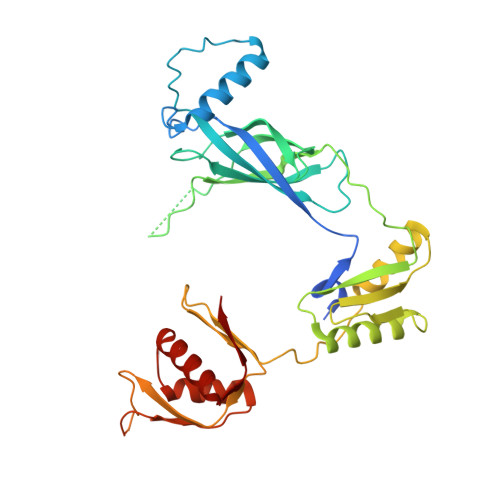
Molecular structure of the intact bacterial flagellar basal body.
Johnson, S., Furlong, E.J., Deme, J.C., Nord, A.L., Caesar, J.J.E., Chevance, F.F.V., Berry, R.M., Hughes, K.T., Lea, S.M.(2021) Nat Microbiol 6: 712-721
- PubMed: 33931760
- DOI: https://doi.org/10.1038/s41564-021-00895-y
- Primary Citation of Related Structures:
7BGL, 7BHQ, 7BIN, 7BJ2, 7BK0, 7NVG - PubMed Abstract:
The bacterial flagellum is a macromolecular protein complex that enables motility in many species. Bacterial flagella self-assemble a strong, multicomponent drive shaft that couples rotation in the inner membrane to the micrometre-long flagellar filament that powers bacterial swimming in viscous fluids 1-3 . Here, we present structures of the intact Salmonella flagellar basal body 4 , encompassing the inner membrane rotor, drive shaft and outer-membrane bushing, solved using cryo-electron microscopy to resolutions of 2.2-3.7 Å. The structures reveal molecular details of how 173 protein molecules of 13 different types assemble into a complex spanning two membranes and a cell wall. The helical drive shaft at one end is intricately interwoven with the rotor component with both the export gate complex and the proximal rod forming interactions with the MS-ring. At the other end, the drive shaft distal rod passes through the LP-ring bushing complex, which functions as a molecular bearing anchored in the outer membrane through interactions with the lipopolysaccharide. The in situ structure of a protein complex capping the drive shaft provides molecular insights into the assembly process of this molecular machine.
Organizational Affiliation:
Sir William Dunn School of Pathology, University of Oxford, Oxford, UK. steven.johnson@path.ox.ac.uk.