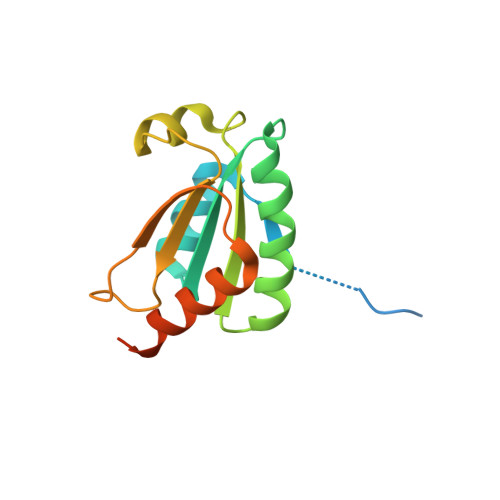
Crystal structure of chloroplastic thioredoxin z defines a type-specific target recognition.
Le Moigne, T., Gurrieri, L., Crozet, P., Marchand, C.H., Zaffagnini, M., Sparla, F., Lemaire, S.D., Henri, J.(2021) Plant J 107: 434-447
- PubMed: 33930214
- DOI: https://doi.org/10.1111/tpj.15300
- Primary Citation of Related Structures:
7ASW - PubMed Abstract:
Thioredoxins (TRXs) are ubiquitous disulfide oxidoreductases structured according to a highly conserved fold. TRXs are involved in a myriad of different processes through a common chemical mechanism. Plant TRXs evolved into seven types with diverse subcellular localization and distinct protein target selectivity. Five TRX types coexist in the chloroplast, with yet scarcely described specificities. We solved the crystal structure of a chloroplastic z-type TRX, revealing a conserved TRX fold with an original electrostatic surface potential surrounding the redox site. This recognition surface is distinct from all other known TRX types from plant and non-plant sources and is exclusively conserved in plant z-type TRXs. We show that this electronegative surface endows thioredoxin z (TRXz) with a capacity to activate the photosynthetic Calvin-Benson cycle enzyme phosphoribulokinase. The distinct electronegative surface of TRXz thereby extends the repertoire of TRX-target recognitions.
Organizational Affiliation:
Laboratoire de Biologie Computationnelle et Quantitative, Institut de Biologie Paris-Seine, UMR 7238, CNRS, Sorbonne Université, 4 Place Jussieu, Paris, 75005, France.