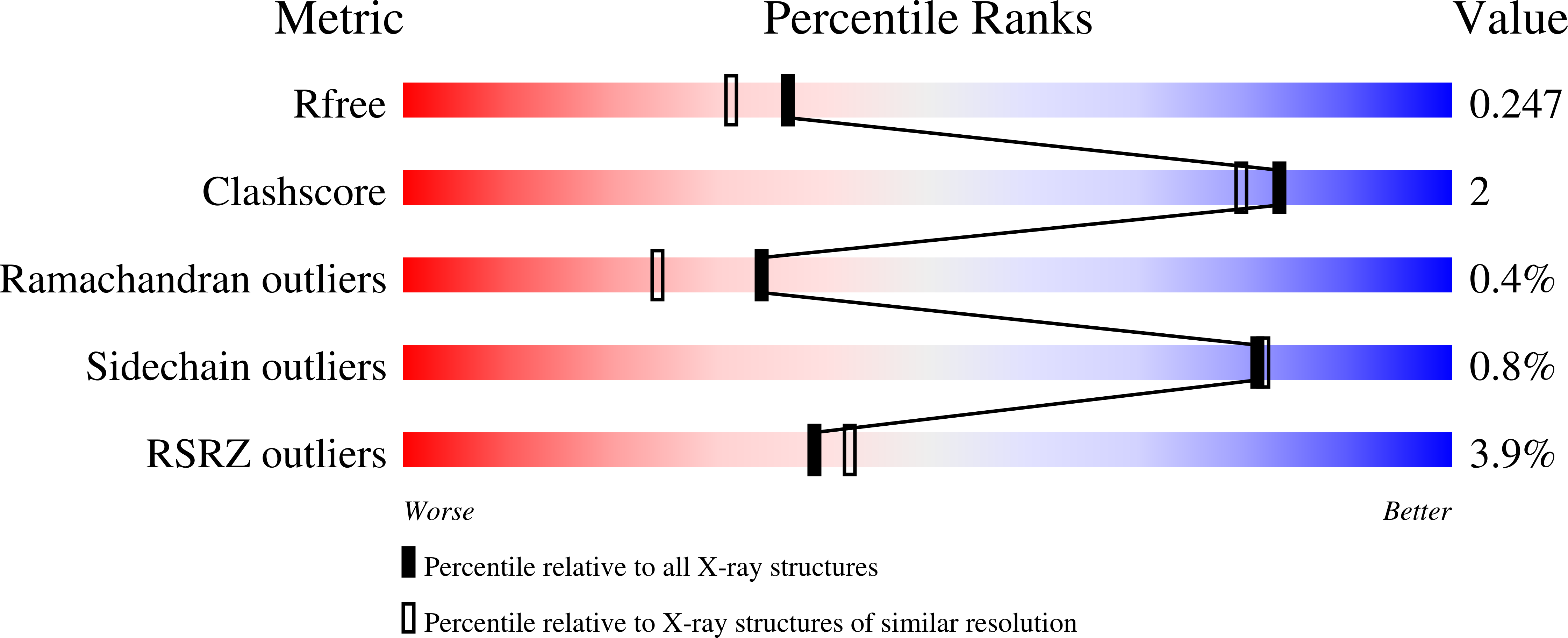
In Vitro and In Vivo Efficacy of NITD-916 against Mycobacterium fortuitum.
Roquet-Baneres, F., Alcaraz, M., Hamela, C., Abendroth, J., Edwards, T.E., Kremer, L.(2023) Antimicrob Agents Chemother 67: e0160722-e0160722
- PubMed: 36920188
- DOI: https://doi.org/10.1128/aac.01607-22
- Primary Citation of Related Structures:
7K73, 7U0O - PubMed Abstract:
Mycobacterium fortuitum represents one of the most clinically relevant rapid-growing mycobacterial species. Treatments are complex due to antibiotic resistance and to severe side effects of effective drugs, prolonged time of treatment, and co-infection with other pathogens. Herein, we explored the activity of NITD-916, a direct inhibitor of the enoyl-ACP reductase InhA of the type II fatty acid synthase in Mycobacterium tuberculosis. We found that this compound displayed very low MIC values against a panel of M. fortuitum clinical strains and exerted potent antimicrobial activity against M. fortuitum in macrophages. Remarkably, the compound was also highly efficacious in a zebrafish model of infection. Short duration treatments were sufficient to significantly protect the infected larvae from M. fortuitum-induced killing, which correlated with reduced bacterial burdens and abscesses. Biochemical analyses demonstrated an inhibition of de novo synthesis of mycolic acids. Resolving the crystal structure of the InhA MFO in complex with NAD and NITD-916 confirmed that NITD-916 is a direct inhibitor of InhA MFO . Importantly, single nucleotide polymorphism leading to a G96S substitution in InhA MFO conferred high resistance levels to NITD-916, thus resolving its target in M. fortuitum. Overall, these findings indicate that NITD-916 is highly active against M. fortuitum both in vitro and in vivo and should be considered in future preclinical evaluations for the treatment of M. fortuitum pulmonary diseases.
Organizational Affiliation:
Centre National de la Recherche Scientifique UMR 9004, Institut de Recherche en Infectiologie de Montpellier (IRIM), Université de Montpellier, Montpellier, France.