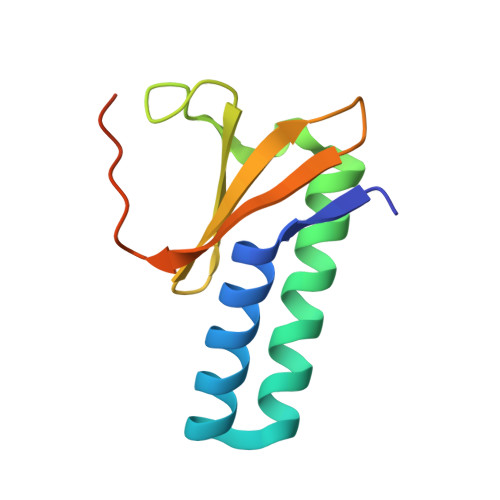
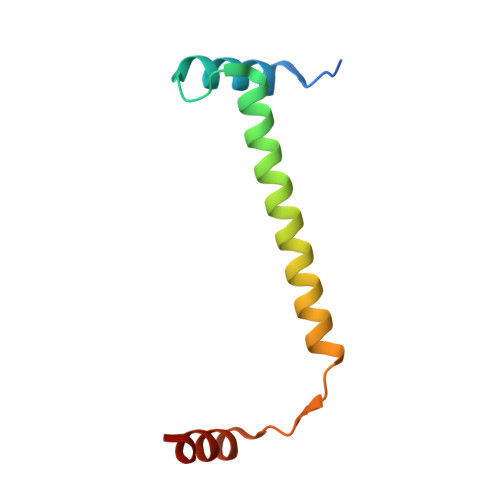
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Garcia-Rodriguez, G., Girardin, Y., Kumar Singh, R., Volkov, A.N., Van Dyck, J., Muruganandam, G., Sobott, F., Charlier, D., Loris, R.(2024) Sci Adv 10: eadj2403-eadj2403
- PubMed: 38181072
- DOI: https://doi.org/10.1126/sciadv.adj2403
- Primary Citation of Related Structures:
7R5A - PubMed Abstract:
The parDE family of toxin-antitoxin (TA) operons is ubiquitous in bacterial genomes and, in Vibrio cholerae , is an essential component to maintain the presence of chromosome II. Here, we show that transcription of the V. cholerae parDE2 (Vc parDE ) operon is regulated in a toxin:antitoxin ratio-dependent manner using a molecular mechanism distinct from other type II TA systems. The repressor of the operon is identified as an assembly with a 6:2 stoichiometry with three interacting ParD2 dimers bridged by two ParE2 monomers. This assembly docks to a three-site operator containing 5'- GGTA-3' motifs. Saturation of this TA complex with ParE2 toxin results in disruption of the interface between ParD2 dimers and the formation of a TA complex of 2:2 stoichiometry. The latter is operator binding-incompetent as it is incompatible with the required spacing of the ParD2 dimers on the operator.
Organizational Affiliation:
Structural Biology Brussels, Department of Biotechnology, Vrije Universiteit Brussel, Pleinlaan 2, B-1050 Brussel, Belgium.