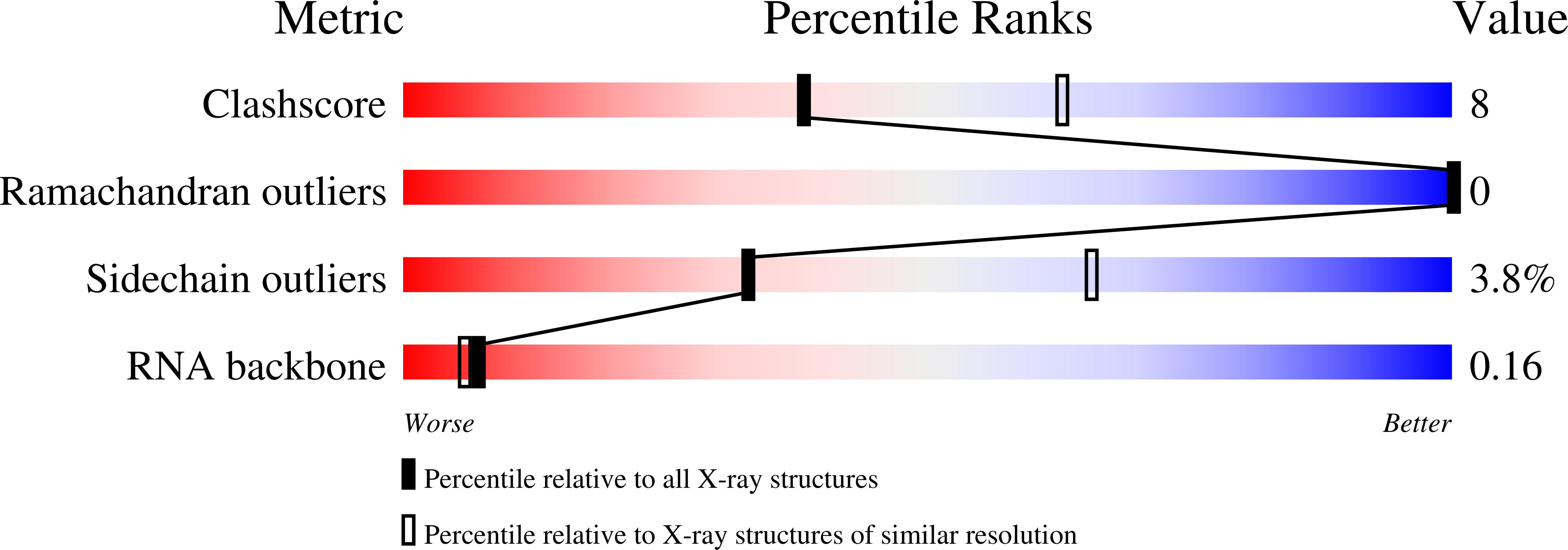
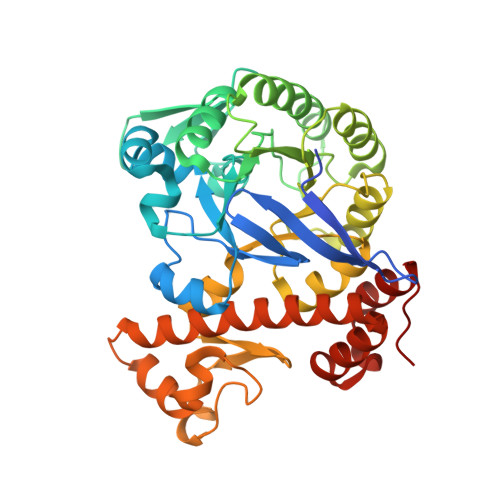
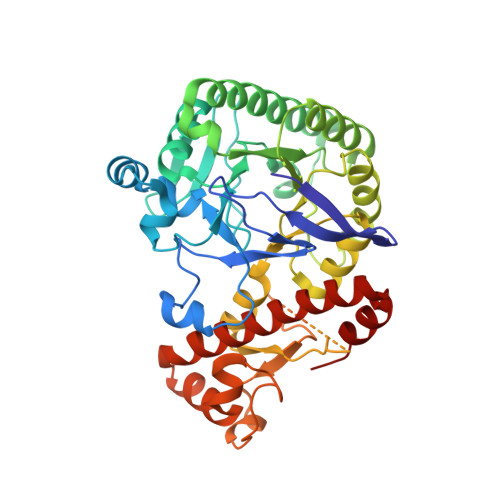
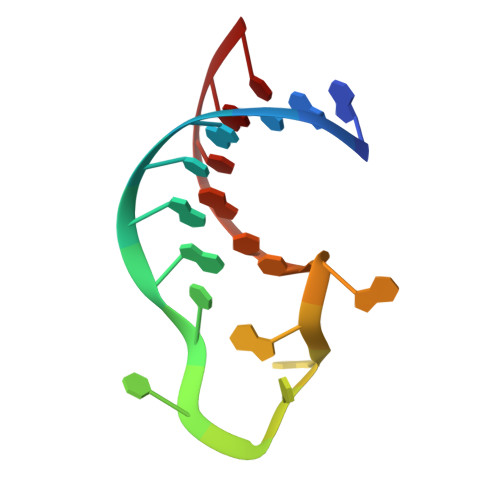
Structural and functional insights into human tRNA guanine transgylcosylase.
Sievers, K., Welp, L., Urlaub, H., Ficner, R.(2021) RNA Biol 18: 382-396
- PubMed: 34241577
- DOI: https://doi.org/10.1080/15476286.2021.1950980
- Primary Citation of Related Structures:
7NQ4 - PubMed Abstract:
The eukaryotic tRNA guanine transglycosylase (TGT) is an RNA modifying enzyme incorporating queuine, a hypermodified guanine derivative, into the tRNAs Asp,Asn,His,Tyr . While both subunits of the functional heterodimer have been crystallized individually, much of our understanding of its dimer interface or recognition of a target RNA has been inferred from its more thoroughly studied bacterial homolog. However, since bacterial TGT, by incorporating queuine precursor preQ 1 , deviates not only in function, but as a homodimer, also in its subunit architecture, any inferences regarding the subunit association of the eukaryotic heterodimer or the significance of its unique catalytically inactive subunit are based on unstable footing. Here, we report the crystal structure of human TGT in its heterodimeric form and in complex with a 25-mer stem loop RNA, enabling detailed analysis of its dimer interface and interaction with a minimal substrate RNA. Based on a model of bound tRNA, we addressed a potential functional role of the catalytically inactive subunit QTRT2 by UV-crosslinking and mutagenesis experiments, identifying the two-stranded βEβF-sheet of the QTRT2 subunit as an additional RNA-binding motif.
Organizational Affiliation:
Department of Molecular Structural Biology, University of Göttingen, Göttingen, Germany.