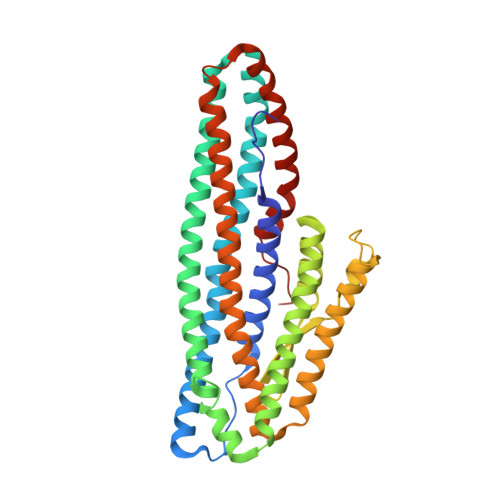
The Crystal Structure of Bacillus cereus HblL 1 .
Worthy, H.L., Williamson, L.J., Auhim, H.S., Leppla, S.H., Sastalla, I., Jones, D.D., Rizkallah, P.J., Berry, C.(2021) Toxins (Basel) 13
- PubMed: 33807365
- DOI: https://doi.org/10.3390/toxins13040253
- Primary Citation of Related Structures:
7NMQ - PubMed Abstract:
The Hbl toxin is a three-component haemolytic complex produced by Bacillus cereus sensu lato strains and implicated as a cause of diarrhoea in B. cereus food poisoning. While the structure of the HblB component of this toxin is known, the structures of the other components are unresolved. Here, we describe the expression of the recombinant HblL 1 component and the elucidation of its structure to 1.36 Å. Like HblB, it is a member of the alpha-helical pore-forming toxin family. In comparison to other members of this group, it has an extended hydrophobic beta tongue region that may be involved in pore formation. Molecular docking was used to predict possible interactions between HblL 1 and HblB, and suggests a head to tail dimer might form, burying the HblL 1 beta tongue region.
Organizational Affiliation:
School of Biosciences, Cardiff University, Park Place, Cardiff CF10 3AX, UK.