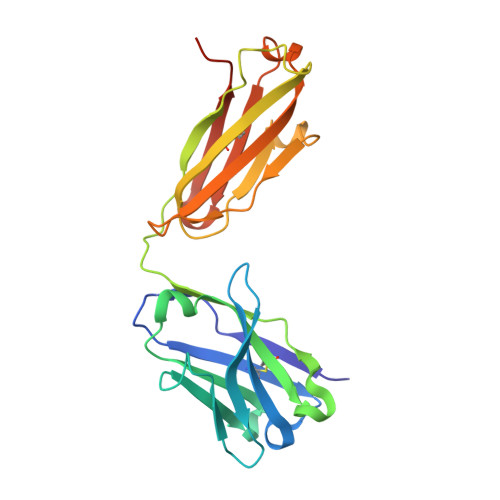
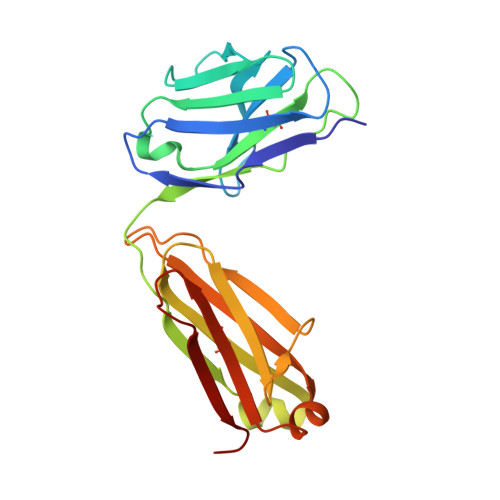
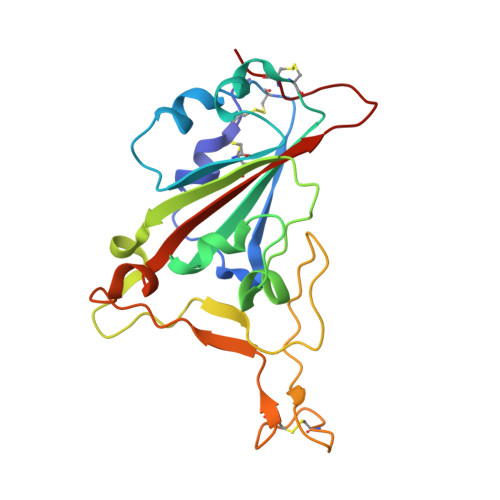
A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes.
Guo, Y., Huang, L., Zhang, G., Yao, Y., Zhou, H., Shen, S., Shen, B., Li, B., Li, X., Zhang, Q., Chen, M., Chen, D., Wu, J., Fu, D., Zeng, X., Feng, M., Pi, C., Wang, Y., Zhou, X., Lu, M., Li, Y., Fang, Y., Lu, Y.Y., Hu, X., Wang, S., Zhang, W., Gao, G., Adrian, F., Wang, Q., Yu, F., Peng, Y., Gabibov, A.G., Min, J., Wang, Y., Huang, H., Stepanov, A., Zhang, W., Cai, Y., Liu, J., Yuan, Z., Zhang, C., Lou, Z., Deng, F., Zhang, H., Shan, C., Schweizer, L., Sun, K., Rao, Z.(2021) Nat Commun 12: 2623-2623
- PubMed: 33976198
- DOI: https://doi.org/10.1038/s41467-021-22926-2
- Primary Citation of Related Structures:
7CJF - PubMed Abstract:
COVID-19 pandemic caused by SARS-CoV-2 constitutes a global public health crisis with enormous economic consequences. Monoclonal antibodies against SARS-CoV-2 can provide an important treatment option to fight COVID-19, especially for the most vulnerable populations. In this work, potent antibodies binding to SARS-CoV-2 Spike protein were identified from COVID-19 convalescent patients. Among them, P4A1 interacts directly with and covers majority of the Receptor Binding Motif of the Spike Receptor-Binding Domain, shown by high-resolution complex structure analysis. We further demonstrate the binding and neutralizing activities of P4A1 against wild type and mutant Spike proteins or pseudoviruses. P4A1 was subsequently engineered to reduce the potential risk for Antibody-Dependent Enhancement of infection and to extend its half-life. The engineered antibody exhibits an optimized pharmacokinetic and safety profile, and it results in complete viral clearance in a rhesus monkey model of COVID-19 following a single injection. These data suggest its potential against SARS-CoV-2 related diseases.
Organizational Affiliation:
State Key Laboratory of Medicinal Chemical Biology and College of Pharmacy, Nankai University, Tianjin, People's Republic of China. guoyu@nankai.edu.cn.