7BZU
Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 5.5
- PDB DOI: https://doi.org/10.2210/pdb7BZU/pdb
- EM Map EMD-30260: EMDB EMDataResource
- Classification: VIRUS
- Organism(s): Coxsackievirus A10, Homo sapiens
- Expression System: Homo sapiens
- Mutation(s): No
- Deposited: 2020-04-28 Released: 2020-07-22
- Funding Organization(s): Chinese Academy of Sciences
Experimental Data Snapshot
- Method: ELECTRON MICROSCOPY
- Resolution: 3.00 Å
- Aggregation State: PARTICLE
- Reconstruction Method: SINGLE PARTICLE
wwPDB Validation 3D Report Full Report
This is version 1.3 of the entry. See complete history.
Macromolecules
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Capsid protein VP1 | 298 | Coxsackievirus A10 | Mutation(s): 0 | 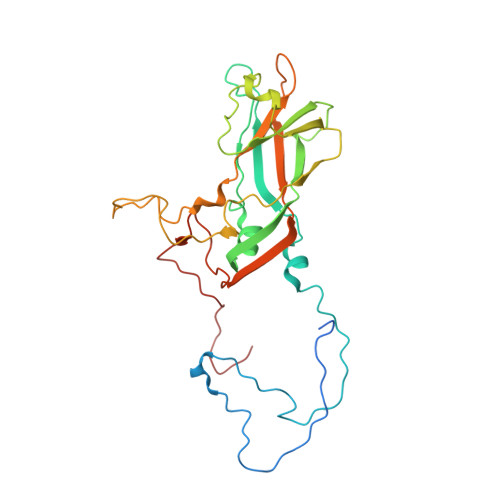 | |
UniProt | |||||
Find proteins for A0A0C5AWF6 (Coxsackievirus A10) Explore A0A0C5AWF6 Go to UniProtKB: A0A0C5AWF6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0C5AWF6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Capsid protein VP2 | 255 | Coxsackievirus A10 | Mutation(s): 0 | 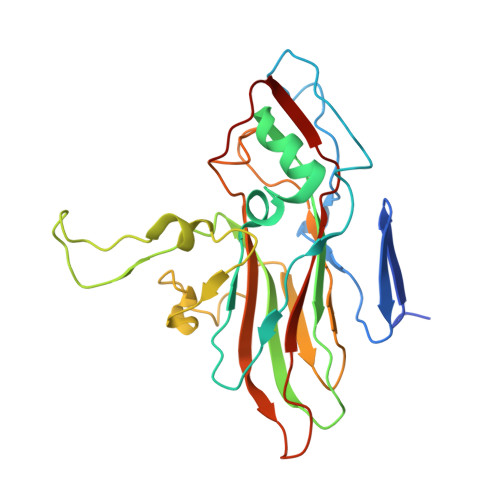 | |
UniProt | |||||
Find proteins for G0YPI2 (Coxsackievirus A10) Explore G0YPI2 Go to UniProtKB: G0YPI2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | G0YPI2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Capsid protein VP3 | 240 | Coxsackievirus A10 | Mutation(s): 0 | 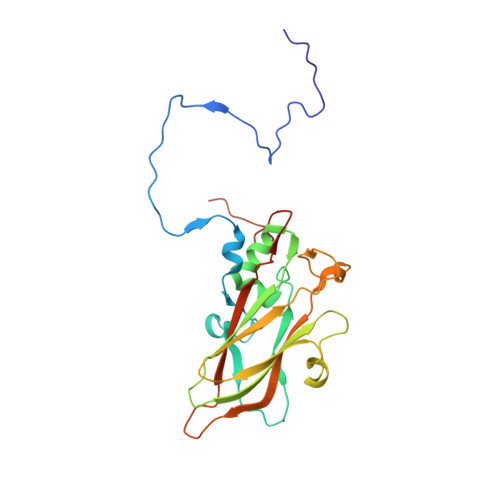 | |
UniProt | |||||
Find proteins for G0YPI2 (Coxsackievirus A10) Explore G0YPI2 Go to UniProtKB: G0YPI2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | G0YPI2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Capsid protein VP4 | 69 | Coxsackievirus A10 | Mutation(s): 0 | 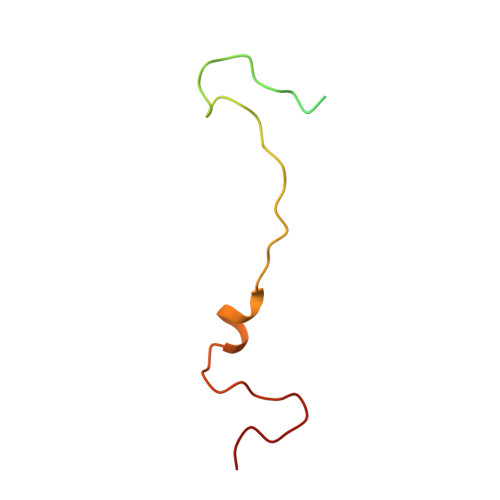 | |
UniProt | |||||
Find proteins for G0YPI2 (Coxsackievirus A10) Explore G0YPI2 Go to UniProtKB: G0YPI2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | G0YPI2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| KRM1 | 375 | Homo sapiens | Mutation(s): 0 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q96MU8 (Homo sapiens) Explore Q96MU8 Go to UniProtKB: Q96MU8 | |||||
PHAROS: Q96MU8 GTEx: ENSG00000183762 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96MU8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Small Molecules
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | F [auth E], G [auth E], H [auth E] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
Experimental Data & Validation
Experimental Data
- Method: ELECTRON MICROSCOPY
- Resolution: 3.00 Å
- Aggregation State: PARTICLE
- Reconstruction Method: SINGLE PARTICLE
Entry History & Funding Information
Deposition Data
- Released Date: 2020-07-22 Deposition Author(s): Cui, Y., Peng, R., Song, H., Tong, Z., Gao, G.F., Qi, J.
| Funding Organization | Location | Grant Number |
|---|---|---|
| Chinese Academy of Sciences | China | XDB29010202 |
Revision History (Full details and data files)
- Version 1.0: 2020-07-22
Type: Initial release - Version 1.1: 2020-07-29
Type: Remediation
Reason: Carbohydrate remediation
Changes: Data collection, Derived calculations, Structure summary - Version 1.2: 2020-08-05
Changes: Database references - Version 1.3: 2020-08-19
Changes: Database references, Structure summary













