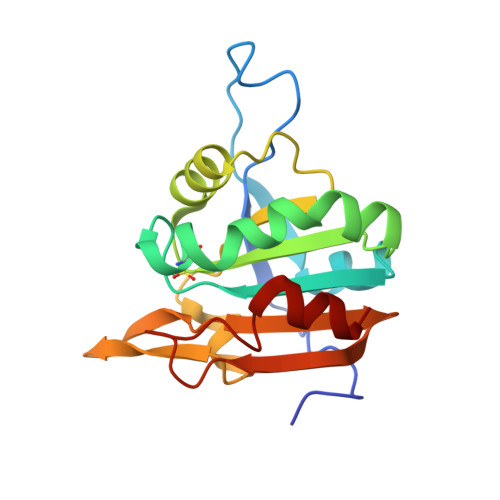
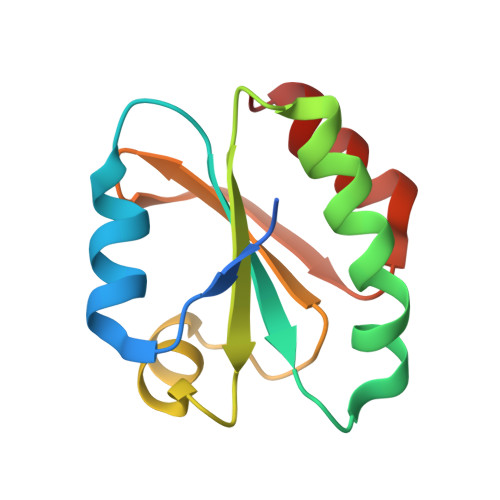
Crystal structure of sulfonic peroxiredoxin Ahp1 in complex with thioredoxin Trx2 mimics a conformational intermediate during the catalytic cycle.
Lian, F.M., Jiang, Y.L., Yang, W., Yang, X.(2020) Int J Biol Macromol 161: 1055-1060
- PubMed: 32531362
- DOI: https://doi.org/10.1016/j.ijbiomac.2020.06.065
- Primary Citation of Related Structures:
7BVV - PubMed Abstract:
Peroxiredoxin (Prx) is a thiol-based peroxidase that eliminates reactive oxygen species to avoid oxidative damage. Alkyl hydroperoxide reductase Ahp1 is a novel and specific typical 2-cysteine Prx. Here, we present the crystal structure of sulfonic Ahp1 complexed with thioredoxin Trx2 at 2.12 Å resolution. This structure implies that the transient Ahp1-Trx2 complex during the catalytic cycle already have an ability to decompose the peroxides. Structural analysis reveals that the segment glutamine23-lysine32 juxtaposed to the resolving cysteine (C R ) of Ahp1 moves inward to generate a compact structure upon peroxidatic cysteine (C P ) overoxidation, resulting in the breakdown of several conserved hydrogen bonds formed by Ahp1-Trx2 complex interaction. Structural comparisons suggest that the structure of sulfonic Ahp1 represents a novel conformation of Ahp1, which can mimic a conformational intermediate between the reduced and oxidized forms. Therefore, this study may provide a new structural insight into the intermediate state in which the segment glutamine23-lysine32 juxtaposed to the cysteine31 (C R ) undergoes a conformational change upon cysteine62 (C P ) oxidation to prepare for the formation of an intermolecular C P -C R disulfide bond during Ahp1 catalytic cycle.
Organizational Affiliation:
Key Laboratory of Precision Oncology of Shandong Higher Education, Institute of Precision Medicine, Jining Medical University, Jining, Shandong 272067, People's Republic of China; Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230027, People's Republic of China. Electronic address: fmlian@mail.jnmc.edu.cn.