Cooperation between peptidoglycan transpeptidases and SEDS proteins in Bacillus subtilis cell division
Sassine, J., Rao, V.A., Goldsmith, G., Breukink, E., Lewis, R.J., Daniel, R.A., Vollmer, W.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Penicillin-binding protein 3 | 662 | Bacillus subtilis subsp. subtilis str. 168 | Mutation(s): 0 Gene Names: pbpC, ycsM, yzsA, BSU04140 EC: 3.4.16.4 | 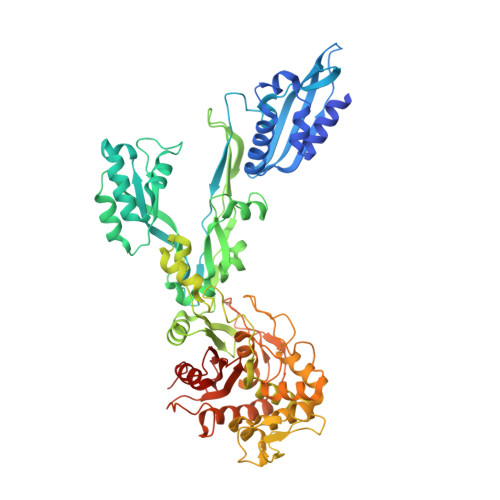 | |
UniProt | |||||
Find proteins for P42971 (Bacillus subtilis (strain 168)) Explore P42971 Go to UniProtKB: P42971 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P42971 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 54.624 | α = 90 |
| b = 108.684 | β = 90 |
| c = 238.691 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| PHENIX | refinement |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Medical Research Council (MRC, United Kingdom) | United Kingdom | MR/N002679/1 |