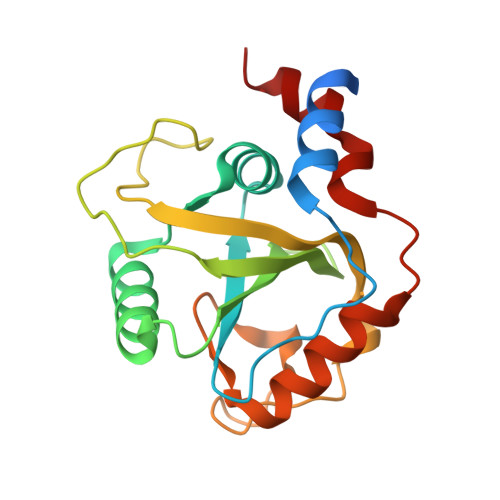
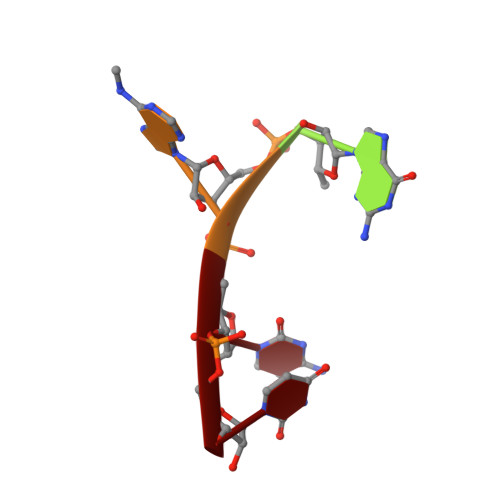
Structural and Dynamic Insights into Redundant Function of YTHDF Proteins.
Li, Y., Bedi, R.K., Moroz-Omori, E.V., Caflisch, A.(2020) J Chem Inf Model 60: 5932-5935
- PubMed: 33073985
- DOI: https://doi.org/10.1021/acs.jcim.0c01029
- Primary Citation of Related Structures:
6ZOT - PubMed Abstract:
Three YTH-domain family proteins (YTHDF1, YTHDF2, and YTHDF3) recognize the N 6 -methyladenosine (m 6 A) modification of mRNA in cells. However, the redundancy of their cellular functions has been disputed. We investigate their interactions with m 6 A-containing RNA using X-ray crystallography and molecular dynamics (MD). The new X-ray structures and MD simulations show that the three proteins share identical interactions with the m 6 A-containing RNA and have similar intrinsic plasticity, thus evidencing the redundant roles of the three proteins in cellular functions.
Organizational Affiliation:
Department of Biochemistry, University of Zurich, CH-8057 Zurich, Switzerland.