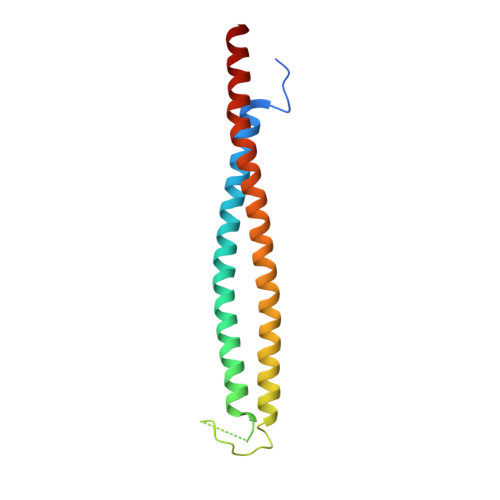
A rod conformation of the Pyrococcus furiosus Rad50 coiled coil.
Soh, Y.M., Basquin, J., Gruber, S.(2021) Proteins 89: 251-255
- PubMed: 32875643
- DOI: https://doi.org/10.1002/prot.26005
- Primary Citation of Related Structures:
6ZFF - PubMed Abstract:
The Rad50-Mre11 nuclease complex plays a vital role in DNA repair in all domains of life. It recognizes and processes DNA double-strand breaks. Rad50 proteins fold into an extended structure with a 20 to 60 nm long coiled coil connecting a globular ABC ATPase domain with a zinc hook dimerization domain. A published structure of an archaeal Rad50 zinc hook shows coiled coils pointing away from each other. Here we present the crystal structure of an alternate conformation displaying co-aligned coiled coils. Archaeal Rad50 may thus switch between rod-shaped and ring-like conformations as recently proposed for a bacterial homolog.
Organizational Affiliation:
Department of Fundamental Microbiology (DMF), Faculty of Biology and Medicine (FBM), University of Lausanne (UNIL), Lausanne, Switzerland.