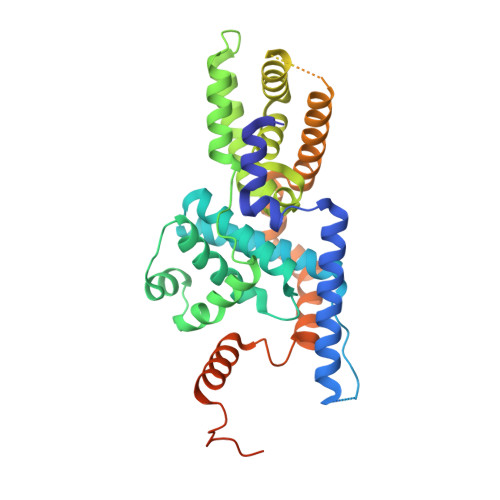
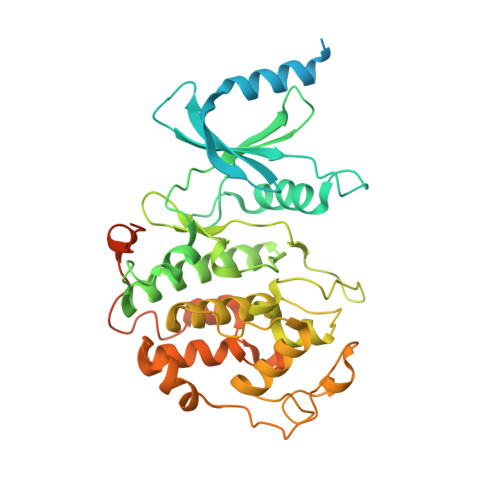
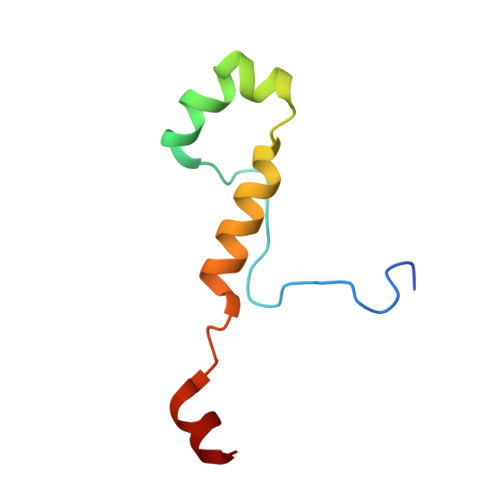
Structural basis for CDK7 activation by MAT1 and Cyclin H.
Peissert, S., Schlosser, A., Kendel, R., Kuper, J., Kisker, C.(2020) Proc Natl Acad Sci U S A 117: 26739-26748
- PubMed: 33055219
- DOI: https://doi.org/10.1073/pnas.2010885117
- Primary Citation of Related Structures:
6Z3U, 6Z4X - PubMed Abstract:
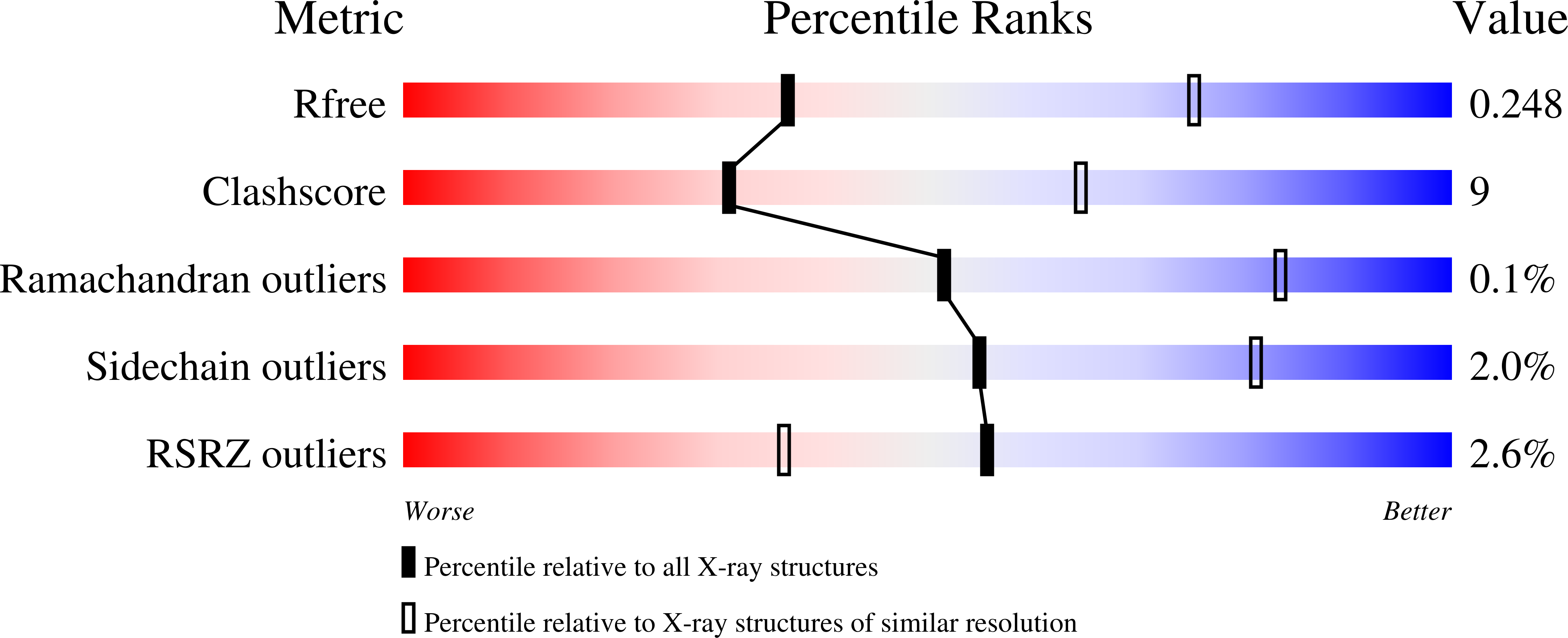
Cyclin-dependent kinase 7 (CDK7), Cyclin H, and the RING-finger protein MAT1 form the heterotrimeric CDK-activating kinase (CAK) complex which is vital for transcription and cell-cycle control. When associated with the general transcription factor II H (TFIIH) it activates RNA polymerase II by hyperphosphorylation of its C-terminal domain (CTD). In the absence of TFIIH the trimeric complex phosphorylates the T-loop of CDKs that control cell-cycle progression. CAK holds a special position among the CDK branch due to this dual activity and the dependence on two proteins for activation. We solved the structure of the CAK complex from the model organism Chaetomium thermophilum at 2.6-Å resolution. Our structure reveals an intricate network of interactions between CDK7 and its two binding partners MAT1 and Cyclin H, providing a structural basis for the mechanism of CDK7 activation and CAK activity regulation. In vitro activity measurements and functional mutagenesis show that CDK7 activation can occur independent of T-loop phosphorylation and is thus exclusively MAT1-dependent by positioning the CDK7 T-loop in its active conformation.
Organizational Affiliation:
Rudolf Virchow Center for Integrative and Translational Bioimaging, Institute for Structural Biology, University of Würzburg, 97080 Würzburg, Germany.